A Systematic Study of Modulation of ADAM-Mediated Ectodomain Shedding by Site-Specific O-Glycosylation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Neprilysin Is Required for Angiotensin-(1-7)
Page 1 of 39 Diabetes NEPRILYSIN IS REQUIRED FOR ANGIOTENSIN-(1-7)’S ABILITY TO ENHANCE INSULIN SECRETION VIA ITS PROTEOLYTIC ACTIVITY TO GENERATE ANGIOTENSIN-(1-2) Gurkirat S. Brara, Breanne M. Barrowa, Matthew Watsonb, Ryan Griesbachc, Edwina Chounga, Andrew Welchc, Bela Ruzsicskad, Daniel P. Raleighb, Sakeneh Zraikaa,c aVeterans Affairs Puget Sound Health Care System, Seattle, WA 98108, United States bDepartment of Chemistry, Stony Brook University, Stony Brook, NY 11794, United States cDivision of Metabolism, Endocrinology and Nutrition, Department of Medicine, University of Washington, Seattle, WA 98195, United States dInstitute for Chemical Biology and Drug Discovery, Stony Brook University, Stony Brook, NY 11794, United States Short Title: Angiotensin-(1-7) and insulin secretion Word count: 3997; Figure count: 8 main (plus 3 Online Suppl.); Table count: 1 Online Suppl. Correspondence to: Sakeneh Zraika, PhD 1660 South Columbian Way (151) Seattle, WA, United States Tel: 206-768-5391 / Fax: 206-764-2164 Email: [email protected] 1 Diabetes Publish Ahead of Print, published online May 30, 2017 Diabetes Page 2 of 39 ABSTRACT Recent work has renewed interest in therapies targeting the renin-angiotensin system (RAS) to improve β-cell function in type 2 diabetes. Studies show that generation of angiotensin-(1-7) by angiotensin converting enzyme 2 (ACE2) and its binding to the Mas receptor (MasR) improves glucose homeostasis, partly by enhancing glucose-stimulated insulin secretion (GSIS). Thus, islet ACE2 upregulation is viewed as a desirable therapeutic goal. Here, we show that although endogenous islet ACE2 expression is sparse, its inhibition abrogates angiotensin-(1-7)-mediated GSIS. However, a more widely expressed islet peptidase, neprilysin, degrades angiotensin-(1-7) into several peptides. -

What Are the Roles of Metalloproteinases in Cartilage and Bone Damage? G Murphy, M H Lee
iv44 Ann Rheum Dis: first published as 10.1136/ard.2005.042465 on 20 October 2005. Downloaded from REPORT What are the roles of metalloproteinases in cartilage and bone damage? G Murphy, M H Lee ............................................................................................................................... Ann Rheum Dis 2005;64:iv44–iv47. doi: 10.1136/ard.2005.042465 enzyme moiety into an upper and a lower subdomain. A A role for metalloproteinases in the pathological destruction common five stranded beta-sheet and two alpha-helices are in diseases such as rheumatoid arthritis and osteoarthritis, always found in the upper subdomain with a further C- and the irreversible nature of the ensuing cartilage and bone terminal helix in the lower subdomain. The catalytic sites of damage, have been the focus of much investigation for the metalloproteinases, especially the MMPs, have been several decades. This has led to the development of broad targeted for the development of low molecular weight spectrum metalloproteinase inhibitors as potential therapeu- synthetic inhibitors with a zinc chelating moiety. Inhibitors tics. More recently it has been appreciated that several able to fully differentiate between individual enzymes have families of zinc dependent proteinases play significant and not been identified thus far, although a reasonable level of varied roles in the biology of the resident cells in these tissues, discrimination is now being achieved in some cases.7 Each orchestrating development, remodelling, and subsequent family does, however, have other unique domains with pathological processes. They also play key roles in the numerous roles, including the determination of physiological activity of inflammatory cells. The task of elucidating the substrate specificity, ECM, or cell surface localisation (fig 1). -

Human ADAM12 Quantikine ELISA
Quantikine® ELISA Human ADAM12 Immunoassay Catalog Number DAD120 For the quantitative determination of A Disintegrin And Metalloproteinase domain- containing protein 12 (ADAM12) concentrations in cell culture supernates, serum, plasma, and urine. This package insert must be read in its entirety before using this product. For research use only. Not for use in diagnostic procedures. TABLE OF CONTENTS SECTION PAGE INTRODUCTION .....................................................................................................................................................................1 PRINCIPLE OF THE ASSAY ...................................................................................................................................................2 LIMITATIONS OF THE PROCEDURE .................................................................................................................................2 TECHNICAL HINTS .................................................................................................................................................................2 MATERIALS PROVIDED & STORAGE CONDITIONS ...................................................................................................3 OTHER SUPPLIES REQUIRED .............................................................................................................................................3 PRECAUTIONS .........................................................................................................................................................................4 -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

3 Cleavage Products of Notch 2/Site and Myelopoiesis by Dysregulating
ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch This information is current as David R. Gibb, Sheinei J. Saleem, Dae-Joong Kang, Mark of October 4, 2021. A. Subler and Daniel H. Conrad J Immunol 2011; 186:4244-4252; Prepublished online 2 March 2011; doi: 10.4049/jimmunol.1003318 http://www.jimmunol.org/content/186/7/4244 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2011/03/02/jimmunol.100331 Material 8.DC1 http://www.jimmunol.org/ References This article cites 45 articles, 16 of which you can access for free at: http://www.jimmunol.org/content/186/7/4244.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 4, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch David R. -

Negatively Regulated by ADAM15 TRIF-Mediated TLR3 and TLR4
TRIF-Mediated TLR3 and TLR4 Signaling Is Negatively Regulated by ADAM15 Suaad Ahmed, Ashwini Maratha, Aisha Qasim Butt, Enda Shevlin and Sinead M. Miggin This information is current as of September 27, 2021. J Immunol 2013; 190:2217-2228; Prepublished online 30 January 2013; doi: 10.4049/jimmunol.1201630 http://www.jimmunol.org/content/190/5/2217 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2013/01/30/jimmunol.120163 Material 0.DC1 References This article cites 57 articles, 20 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/190/5/2217.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 27, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology TRIF-Mediated TLR3 and TLR4 Signaling Is Negatively Regulated by ADAM15 Suaad Ahmed, Ashwini Maratha, Aisha Qasim Butt, Enda Shevlin, and Sinead M. -

ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease
International Journal of Molecular Sciences Review ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease Francesca Tosetti 1,* , Massimo Alessio 2, Alessandro Poggi 1,† and Maria Raffaella Zocchi 3,† 1 Molecular Oncology and Angiogenesis Unit, IRCCS Ospedale Policlinico S. Martino Largo R. Benzi 10, 16132 Genoa, Italy; [email protected] 2 Proteome Biochemistry, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] 3 Division of Immunology, Transplants and Infectious Diseases, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] * Correspondence: [email protected] † These authors contributed equally to this work as last author. Abstract: Enzymes, once considered static molecular machines acting in defined spatial patterns and sites of action, move to different intra- and extracellular locations, changing their function. This topological regulation revealed a close cross-talk between proteases and signaling events involving post-translational modifications, membrane tyrosine kinase receptors and G-protein coupled recep- tors, motor proteins shuttling cargos in intracellular vesicles, and small-molecule messengers. Here, we highlight recent advances in our knowledge of regulation and function of A Disintegrin And Metalloproteinase (ADAM) endopeptidases at specific subcellular sites, or in multimolecular com- plexes, with a special focus on ADAM10, and tumor necrosis factor-α convertase (TACE/ADAM17), since these two enzymes belong to the same family, share selected substrates and bioactivity. We will discuss some examples of ADAM10 activity modulated by changing partners and subcellular compartmentalization, with the underlying hypothesis that restraining protease activity by spatial Citation: Tosetti, F.; Alessio, M.; segregation is a complex and powerful regulatory tool. -

Cell-Autonomous FLT3L Shedding Via ADAM10 Mediates Conventional Dendritic Cell Development in Mouse Spleen
Cell-autonomous FLT3L shedding via ADAM10 mediates conventional dendritic cell development in mouse spleen Kohei Fujitaa,b,1, Svetoslav Chakarovc,1, Tetsuro Kobayashid, Keiko Sakamotod, Benjamin Voisind, Kaibo Duanc, Taneaki Nakagawaa, Keisuke Horiuchie, Masayuki Amagaib, Florent Ginhouxc, and Keisuke Nagaod,2 aDepartment of Dentistry and Oral Surgery, Keio University School of Medicine, Tokyo 160-8582, Japan; bDepartment of Dermatology, Keio University School of Medicine, Tokyo 160-8582, Japan; cSingapore Immunology Network, Agency for Science, Technology and Research, Biopolis, 138648 Singapore; dDermatology Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892; and eDepartment of Orthopedic Surgery, National Defense Medical College, Tokorozawa 359-8513, Japan Edited by Kenneth M. Murphy, Washington University School of Medicine, St. Louis, MO, and approved June 10, 2019 (received for review November 4, 2018) Conventional dendritic cells (cDCs) derive from bone marrow (BM) intocDC1sorcDC2stakesplaceintheBM(3),andthese precursors that undergo cascades of developmental programs to pre-cDC1s and pre-cDC2s ultimately differentiate into cDC1s terminally differentiate in peripheral tissues. Pre-cDC1s and pre- and cDC2s after migrating to nonlymphoid and lymphoid tissues. + + cDC2s commit in the BM to each differentiate into CD8α /CD103 cDCs are short-lived, and their homeostatic maintenance relies + cDC1s and CD11b cDC2s, respectively. Although both cDCs rely on on constant replenishment from the BM precursors (5). The cy- the cytokine FLT3L during development, mechanisms that ensure tokine Fms-related tyrosine kinase 3 ligand (FLT3L) (12), by cDC accessibility to FLT3L have yet to be elucidated. Here, we gen- signaling through its receptor FLT3 expressed on DC precursors, erated mice that lacked a disintegrin and metalloproteinase (ADAM) is essential during the development of DCs (7, 13). -

Expression of the Disintegrin Metalloprotease, ADAM-10, In
314 Vol. 10, 314–323, January 1, 2004 Clinical Cancer Research Expression of the Disintegrin Metalloprotease, ADAM-10, in Prostate Cancer and Its Regulation by Dihydrotestosterone, Insulin-Like Growth Factor I, and Epidermal Growth Factor in the Prostate Cancer Cell Model LNCaP Daniel R. McCulloch,1 Pascal Akl,1 Conclusions: This study describes for the first time Hemamali Samaratunga,2 Adrian C. Herington,1 the expression, regulation, and cellular localization of and Dimitri M. Odorico1 ADAM-10 protein in PCa. The regulation and membrane 1 localization of ADAM-10 support our hypothesis that Hormone-Dependent Cancer Program, School of Life Sciences, ADAM-10 has a role in extracellular matrix maintenance Queensland University of Technology, Brisbane, Queensland, Australia, and 2Sullivan Nicolaides Pathology, Brisbane, Queensland, and cell invasion, although the potential role of nuclear Australia ADAM-10 is not yet known. INTRODUCTION ABSTRACT The disintegrin metalloproteases ADAMs, like the matrix Purpose: The disintegrin metalloprotease ADAM-10 is metalloproteinases (MMPs), are members of the metzincin a multidomain metalloprotease that is potentially significant (zinc-dependent metalloprotease) superfamily. To date, more in tumor progression due to its extracellular matrix-degrad- than 30 ADAMs have been characterized (1), some of which are ing properties. Previously, ADAM-10 mRNA was detected involved in diverse biological functions such as fertilization, in prostate cancer (PCa) cell lines; however, the presence of neurogenesis (2, 3), and the ectodomain shedding of growth ADAM-10 protein and its cellular localization, regulation, factors such as amyloid precursor protein and tumor necrosis and role have yet to be described. We hypothesized that factor ␣ (4, 5). -

ADAM9: a Novel Player in Vestibular Schwannoma Pathogenesis
1856 ONCOLOGY LETTERS 19: 1856-1864, 2020 ADAM9: A novel player in vestibular schwannoma pathogenesis MARIA BREUN1, ALEXANDRA SCHWERDTFEGER1, DONATO DANIEL MARTELLOTTA1, ALMUTH F. KESSLER1, CAMELIA M. MONORANU2, CORDULA MATTHIES1, MARIO LÖHR1* and CARSTEN HAGEMANN1* 1Department of Neurosurgery, University Hospital Würzburg; 2Department of Neuropathology, Institute of Pathology, University of Würzburg, D-97080 Würzburg, Germany Received April 27, 2019; Accepted October 2, 2019 DOI: 10.3892/ol.2020.11299 Abstract. A disintegrin and metalloproteinase 9 (ADAM9) is VS samples (n=60). A total of 30 of them were from patients a member of the transmembrane ADAM family. It is expressed with neurofibromatosis. Healthy peripheral nerves from in different types of solid cancer and promotes tumor invasive- autopsies (n=10) served as controls. ADAM9 mRNA levels ness. To the best of our knowledge, the present study was the were measured by PCR, and protein levels were determined first to examine ADAM9 expression in vestibular schwan- by immunohistochemistry (IHC) and western blotting (WB). nomas (VS) from patients with and without neurofibromatosis The Hannover Classification was used to categorize tumor type 2 (NF2) and to associate the data with clinical parameters extension and hearing loss. ADAM9 mRNA levels were of the patients. The aim of the present study was to evaluate 8.8-fold higher in VS compared with in controls. The levels if ADAM9 could be used as prognostic marker or therapeutic were 5.6-fold higher in patients with NF2 and 12-fold higher in target. ADAM9 mRNA and protein levels were measured in patients with sporadic VS. WB revealed two mature isoforms of the protein, and according to IHC ADAM9 was mainly expressed by S100-positive Schwann cells. -
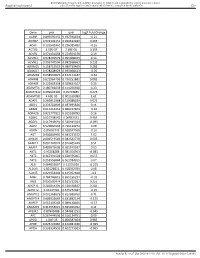
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

Importance of Β-APP, ADAM9, 10, 17 in Alzheimer Disease
Available online at www.ijmrhs.com al R edic ese M a of rc l h a & n r H u e o a J l l t h International Journal of Medical Research & a n S ISSN No: 2319-5886 o c i t i Health Sciences, 2019, 8(7): 68-7 e a n n c r e e t s n I • • I J M R H S Importance of β-APP, ADAM9, 10, 17 in Alzheimer Disease: Preliminary Autopsy Study with Immunohistochemical Expression in Human Brain Filiz Eren1, Nursel Turkmen Inanır1,2, Recep Fedakar2, Bulent Eren3*, Murat Serdar Gurses1, Mustafa Numan Ural4, Sumeyya Akyol5, Busra Aynekin5 and Kadir Demircan5 1 Bursa Morgue Department, Council of Forensic Medicine of Turkey, Bursa, Turkey 2 Department of Forensic Medicine, School of Medicine, Uludag University, Bursa, Turkey 5 3 Department of Forensic Medicine, School of Medicine, Tokat Gaziosmanpaşa University Tokat, Turkey 4 5 Bilgemed Work Safety and Security, Bursa, Turkey 5 Independent Researcher, Ankara, Turkey *Corresponding e-mail: [email protected] ABSTRACT Alzheimer’s disease (AD) is encountered as an important health problem. It was exposed that in the pathophysiology of AD, formation, and aggregation of amyloid β from amyloid precursor protein ( APP), was restrained by α-secretase group, ADAM (a disintegrin and metalloproteinase) enzymes. From this perspective, ADAM group of enzymes can be presumably used in the future both as a diagnostic marker, and potential treatment modality. In our study, 9 cases with or without AD in different age groups with various causes of death who were autopsied in the Bursa Morgue Department of the Council of Forensic Medicine of Turkey were included in the study.