Accessory Publication
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-Like Mouse Models: Tracking the Role of the Hairless Gene
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2006 Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene Yutao Liu University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Life Sciences Commons Recommended Citation Liu, Yutao, "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino- like Mouse Models: Tracking the Role of the Hairless Gene. " PhD diss., University of Tennessee, 2006. https://trace.tennessee.edu/utk_graddiss/1824 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Yutao Liu entitled "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the requirements for the degree of Doctor of Philosophy, with a major in Life Sciences. Brynn H. Voy, Major Professor We have read this dissertation and recommend its acceptance: Naima Moustaid-Moussa, Yisong Wang, Rogert Hettich Accepted for the Council: Carolyn R. -

Mass Spectrometry-Based Proteomics Techniques and Their Application in Ovarian Cancer Research Agata Swiatly, Szymon Plewa, Jan Matysiak and Zenon J
Swiatly et al. Journal of Ovarian Research (2018) 11:88 https://doi.org/10.1186/s13048-018-0460-6 REVIEW Open Access Mass spectrometry-based proteomics techniques and their application in ovarian cancer research Agata Swiatly, Szymon Plewa, Jan Matysiak and Zenon J. Kokot* Abstract Ovarian cancer has emerged as one of the leading cause of gynecological malignancies. So far, the measurement of CA125 and HE4 concentrations in blood and transvaginal ultrasound examination are essential ovarian cancer diagnostic methods. However, their sensitivity and specificity are still not sufficient to detect disease at the early stage. Moreover, applied treatment may appear to be ineffective due to drug-resistance. Because of a high mortality rate of ovarian cancer, there is a pressing need to develop innovative strategies leading to a full understanding of complicated molecular pathways related to cancerogenesis. Recent studies have shown the great potential of clinical proteomics in the characterization of many diseases, including ovarian cancer. Therefore, in this review, we summarized achievements of proteomics in ovarian cancer management. Since the development of mass spectrometry has caused a breakthrough in systems biology, we decided to focus on studies based on this technique. According to PubMed engine, in the years 2008–2010 the number of studies concerning OC proteomics was increasing, and since 2010 it has reached a plateau. Proteomics as a rapidly evolving branch of science may be essential in novel biomarkers discovery, therapy decisions, progression predication, monitoring of drug response or resistance. Despite the fact that proteomics has many to offer, we also discussed some limitations occur in ovarian cancer studies. -

Studies on the Proteome of Human Hair - Identifcation of Histones and Deamidated Keratins Received: 15 August 2017 Sunil S
www.nature.com/scientificreports OPEN Studies on the Proteome of Human Hair - Identifcation of Histones and Deamidated Keratins Received: 15 August 2017 Sunil S. Adav 1, Roopa S. Subbaiaih2, Swat Kim Kerk 2, Amelia Yilin Lee 2,3, Hui Ying Lai3,4, Accepted: 12 January 2018 Kee Woei Ng3,4,7, Siu Kwan Sze 1 & Artur Schmidtchen2,5,6 Published: xx xx xxxx Human hair is laminar-fbrous tissue and an evolutionarily old keratinization product of follicle trichocytes. Studies on the hair proteome can give new insights into hair function and lead to the development of novel biomarkers for hair in health and disease. Human hair proteins were extracted by detergent and detergent-free techniques. We adopted a shotgun proteomics approach, which demonstrated a large extractability and variety of hair proteins after detergent extraction. We found an enrichment of keratin, keratin-associated proteins (KAPs), and intermediate flament proteins, which were part of protein networks associated with response to stress, innate immunity, epidermis development, and the hair cycle. Our analysis also revealed a signifcant deamidation of keratin type I and II, and KAPs. The hair shafts were found to contain several types of histones, which are well known to exert antimicrobial activity. Analysis of the hair proteome, particularly its composition, protein abundances, deamidated hair proteins, and modifcation sites, may ofer a novel approach to explore potential biomarkers of hair health quality, hair diseases, and aging. Hair is an important and evolutionarily conserved structure. It originates from hair follicles deep within the der- mis and is mainly composed of hair keratins and KAPs, which form a complex network that contributes to the rigidity and mechanical properties. -

Table SD1. Patient Characteristicsa
Table SD1. Patient characteristicsa Patient Sex Age Esophageal Treatment Maximum Cell Maximum Maximum Genotype Food SPT/Ab SPT/F b RAST b Rhinitisc Atopicc Asthmac Alternative diagnosis Dated (year) Disease eosinophils thickness in mast cells lymphocytes anaphylaxis (positive dermatitis /hpf basal layer /hpf /hpf reaction) 1 M 11 NL None 0 3 5 3 Unk No ND ND ND Yes No No Recurrent croup December 2 M 11 NL LTRA 0 3 4 3 Unk No ND ND ND No No Yes Functional abdominal pain May 3 F 9 NL None 0 3 4 3 Unk Unk ND ND ND Unk Unk Unk Functional abdominal pain March 4 M 14 NL None 0 2 6 4 Unk No ND ND ND No No No Vomiting/diarrhea Febuary 5 F 7 NL LTRA 0 3 5 6 Unk Yes 1 3 ND Unk Unk Yes Functional abdominal pain March 6 F 13 NL None 0 2 4 2 Unk No 0 4 ND No No No Functional abdominal pain August 7 M 17 CE PPI 0 4 7 12 Unk Yes 17 4 ND No No Yes None November 8 M 6 CE PPI 0 4 6 10 Unk Unk ND ND ND Unk Unk Unk None June 9 F 16 CE LTRA 3 4 6 8 Unk No ND ND ND No No Yes None January 10 F 13 CE LTRA+PPI 3 5 4 8 Unk No 0 0 ND No No No None August 11 F 11 CE LTRA+PPI 6 4 6 9 Unk Unk ND ND ND Unk Unk Unk None May 12 M 11 EE PPI 24 6 6 12 TT No 2 5 3 Yes No Yes None November 13 F 4 EE PPI 25 6 15 15 TT No 0 0 ND No No No None November 14 M 15 EE None 30 6 24 6 TG Unk 1 1 ND Unk Unk Yes None February 15 M 15 EE None 31 6 15 11 TT No 8 2 ND Yes No Yes None March 16 M 13 EE PPI 32 6 10 25 TG No 0 0 ND No No No None June 17 M 6 EE PPI 40 7 10 21 TT Yes 5 3 0 Unk Unk No None November 18 M 13 EE LTRA 42 7 10 5 TT No 4 5 2 Yes Yes Yes None November 19 F 16 EE LTRA+PPI -
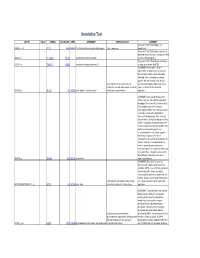
Supplementary Table 5. Functional Annotation of the Largest Gene Cluster(221 Element)
Annotation Tool AFFYID VALUE SYMBOL LOCUSLINK OMIM GENENAME GENEONTOLOGY SUMMARY [Proteome FUNCTION:] Expressed 203054_s_at TCTA 6988 600690 T-cell leukemia translocation altered gene tumor suppressor ubiquitously [Proteome FUNCTION:] May be involved in protein-protein interactions; contains five WD 44563_at FLJ10385 55135 hypothetical protein FLJ10385 domains (WD-40 repeats) [Proteome FUNCTION:] Weakly similarity to 212261_at TNRC15 26058 trinucleotide repeat containing 15 a region of rat nestin (Rn.9701) [SUMMARY:] Actin alpha 1 which is expressed in skeletal muscle is one of six different actin isoforms which have been identified. Actins are highly conserved proteins that are involved in cell motility, actin filament; motor activity; muscle structure and integrity. Alpha actins are a contraction; muscle development; structural major constituent of the contractile 203872_at ACTA1 58 102610 actin, alpha 1, skeletal muscle constituent of cytoskeleton apparatus. [SUMMARY:] Annexin VIII belong to the family of Ca (2+) dependent phospholipid binding proteins (annexins), and has a high 56% identity to annexin V (vascular anticoagulant-alpha). It was initially isolated as 2.2 kb vascular anticoagulant-beta transcript from human placenta, a Ca (2+) dependent phospholipid binding protein that inhibits coagulation and phospholipase A2 activity. However, the fact that annexin VIII is neither an extracellular protein nor associated with the cell surface suggests that it may not play a role in blood coagulation in vivo and its physiological role remains unknown. It is expressed at low levels in human placenta and shows restricted expression in lung endothelia, skin, liver, and kidney. The gene is also found to be selectively overexpressed in acute 203074_at ANXA8 244 602396 annexin A8 myelocytic leukemia. -

Clinical Chemistry
June 1 2007, Volume 53, Issue 6,pp.999- 1180 Editorials Andrew McCaddon and Peter R. Hudson Methylation and Phosphorylation: A Tangled Relationship? Clin Chem 2007 53: 999-1000. Bob Palais Quantitative Heteroduplex Analysis Clin Chem 2007 53: 1001-1003. Eleftherios P. Diamandis Oncopeptidomics: A Useful Approach for Cancer Diagnosis? Clin Chem 2007 53: 1004-1006. Roger D. Klein The Pain Protective Haplotype: Introducing the Modern Genetic Test Clin Chem 2007 53: 1007-1009. Molecular Diagnostics and Genetics Jörn Lötsch, Inna Belfer, Anja Kirchhof, Bikash K. Mishra, Mitchell B. Max, Alexandra Doehring, Michael Costigan, Clifford J. Woolf, Gerd Geisslinger, and Irmgard Tegeder Reliable Screening for a Pain-Protective Haplotype in the GTP Cyclohydrolase 1 Gene (GCH1) Through the Use of 3 or Fewer Single Nucleotide Polymorphisms Clin Chem 2007 53: 1010-1015. Published online March 15, 2007; 10.1373/clinchem.2006.082883 Kerstin L. Edlefsen, Jonathan F. Tait, Mark H. Wener, and Michael Astion Utilization and Diagnostic Yield of Neurogenetic Testing at a Tertiary Care Facility Clin Chem 2007 53: 1016-1022. Published online April 19, 2007; 10.1373/clinchem.2006.083360 Sara Bremer, Helge Rootwelt, and Stein Bergan Real-Time PCR Determination of IMPDH1 and IMPDH2 Expression in Blood Cells Clin Chem 2007 53: 1023-1029. Published online April 26, 2007; 10.1373/clinchem.2006.081968 Elizabeth Herness Peters, Sandra Rojas-Caro, Mitchell G. Brigell, Robert J. Zahorchak, Shelley Ann des Etages, Patricia L. Ruppel, Charles R. Knight, Bradley Austermiller, Myrna C. Graham, Steve Wowk, Sean Banks, Lakshmi V. Madabusi, Patrick Turk, Donna Wilder, Carole Kempfer, Terry W. Osborn, and James C. -

Gene Network and Pathway Analysis of Transcriptional Signatures Characterizing Sole Ulcer and Digital Dermatitis in Dairy Cows
South Dakota State University Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange Electronic Theses and Dissertations 2021 Gene Network and Pathway Analysis of Transcriptional Signatures Characterizing Sole Ulcer and Digital Dermatitis in Dairy Cows Roshin Anie Mohan South Dakota State University Follow this and additional works at: https://openprairie.sdstate.edu/etd Part of the Dairy Science Commons Recommended Citation Mohan, Roshin Anie, "Gene Network and Pathway Analysis of Transcriptional Signatures Characterizing Sole Ulcer and Digital Dermatitis in Dairy Cows" (2021). Electronic Theses and Dissertations. 5278. https://openprairie.sdstate.edu/etd/5278 This Dissertation - Open Access is brought to you for free and open access by Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. For more information, please contact [email protected]. GENE NETWORK AND PATHWAY ANALYSIS OF TRANSCRIPTIONAL SIGNATURES CHARACTERIZING SOLE ULCER AND DIGITAL DERMATITIS IN DAIRY COWS BY ROSHIN ANIE MOHAN A dissertation submitted in partial fulfillment of the requirements for the Doctor of Philosophy Major in Biological Sciences Specialization in Dairy Science South Dakota State University 2021 ii DISSERTATION ACCEPTANCE PAGE Roshin Anie Mohan This dissertation is approved as a creditable and independent investigation by a candidate for the Doctor of Philosophy degree and is acceptable for meeting the dissertation requirements for this degree. Acceptance of this does not imply that the conclusions reached by the candidate are necessarily the conclusions of the major department. -

1 Supplementary Table S1. Histological And
Supplementary Table S1. Histological and immunocytochemestry of the bcMCF clones. Clones Tumor type AE1 CAM5.2 EMA Vimentin bcMCF-1 Invasive poorly - - - +++ bcMCF-4 differentiated spindle cell type bcMCF-2 Invasive poorly +/- +/- +/- ++ bcMCF-6 differentiated bcMCF-7 epithelial cell type bcMCF-3 Invasive poorly - +/- +/- ++ bcMCF-5 differentiated with mix features of spindle and epithelial type The mouse monoclonal antibodies anti- human cytokeratin of low molecular weight (AE1, Biogenex, San Ramon, CA), cytokeratin peptides 7 and 8 (CAM5.2, Ventana, Tucson, AZ), epithelial membrane antigen (EMA) clone E29 and vimentin, clone V9, both from DakoCytomation Inc. (Fort Collins, CO) were used. Negative (-), weak (+/-), moderate (++) or strong (+++). 1 Supplementary Table S2. Differentially expressed apoptosis genes (GO:00009165) in tumorigenic bcMCF cells. Fold change Symbol Gene name trMCF bcMC caMCF AHR aryl hydrocarbon receptor Ns* -2.1 Ns APP amyloid beta (A4) precursor protein Ns -1.7 -1.7 BAG1 BCL2-associated athanogene Ns -7.6 -9.2 BAG5 BCL2-associated athanogene 5 Ns -1.8 Ns BIRC4 baculoviral IAP repeat-containing 4 Ns -2.0 -2.7 BNIP3L BCL2/adenovirus E1B interacting protein 3-like Ns -2 -1.9 CASP14 caspase 14, apoptosis-related cysteine peptidase Ns -12 -12.3 CASP3 caspase 3, apoptosis-related cysteine peptidase Ns -2.4 Ns CASP6 caspase 6, apoptosis-related cysteine peptidase Ns -4.1 -4.1 CD14 CD14 antigen Ns -4.7 Ns DAPK1 death-associated protein kinase 1 Ns -4.1 -13.4 ELMO3 engulfment and cell motility 3 Ns -9.9 -10.8 ELMOD2 ELMO -

Table S2.A-B. Differentially Expressed Genes Following Activation of OGR1 by Acidic Ph in Mouse Peritoneal Macrophages Ph 6.7 24 H
Table S2.A-B. Differentially expressed genes following activation of OGR1 by acidic pH in mouse peritoneal macrophages pH 6.7 24 h. A. Gene List, including gene process B. Complete Table (Excel). Rank Symbol Full name Involved in: WT/KO (Reference: Gene Card, NCBI, JAX, Uniprot, Ratio unless otherwise indicated) 1. Cyp11a1 Cholesterol side chain cleavage Cholesterol, lipid or steroid metabolism. enzyme, mitochondrial (Cytochrome Catalyses the side-chain cleavage reaction of P450 11A1) cholesterol to pregnenolone. 2. Sparc Secreted acidic cysteine rich Cell adhesion, wound healing, ECM glycoprotein (Osteonectin, Basement interactions, bone mineralization. Activates membrane protein 40 (BM-40)) production and activity of matrix metalloproteinases. 3. Tpsb2 Tryptase beta-2 or tryptase II (trypsin- Inflammatory response, proteolysis. like serine protease) 4. Inhba Inhibin Beta A or Activin beta-A chain Immune response and mediators of inflammation and tissue repair.2-5 5. Cpe Carboxypeptidase E Insulin processing, proteolysis. 6. Igfbp7 Insulin-like growth factor-binding Stimulates prostacyclin (PGI2) production and protein 7 cell adhesion. Induced by retinoic acid. 7. Clu Clusterin Chaperone-mediated protein folding, positive regulation of NF-κB transcription factor activity. Protects cells against apoptosis and cytolysis by complement. Promotes proteasomal degradation of COMMD1 and IKBKB. 8. Cma1 Chymase 1 Cellular response to glucose stimulus, interleukin-1 beta biosynthetic process. Possible roles: vasoactive peptide generation, extracellular matrix degradation. 9. Sfrp4 Secreted frizzled-related protein 4 Negative regulation of Wnt signalling. Increases apoptosis during ovulation. Phosphaturic effects by specifically inhibiting sodium-dependent phosphate uptake. 10. Ephx2 Bifunctional epoxide hydrolase Cholesterol homeostasis, xenobiotic metabolism by degrading potentially toxic epoxides. -

Human Hair Keratin Hydrogel : Fabrication, Characterization and Application
This document is downloaded from DR‑NTU (https://dr.ntu.edu.sg) Nanyang Technological University, Singapore. Human hair keratin hydrogel : fabrication, characterization and application Wang, Shuai 2014 Wang, S. (2014). Human hair keratin hydrogel : fabrication, characterization and application. Doctoral thesis, Nanyang Technological University, Singapore. https://hdl.handle.net/10356/62055 https://doi.org/10.32657/10356/62055 Downloaded on 04 Oct 2021 06:13:17 SGT HUMAN HAIR KERATIN HYDROGEL: FABRICATION, CHARACTERIZATION AND APPLICATION WANG SHUAI SCHOOL OF MATERIALS SCIENCE AND ENGINEERING 2014 HUMAN HAIR KERATIN HYDROGEL: FABRICATION, CHARACTERIZATION AND APPLICATION WANG SHUAI School of Materials Science and Engineering A thesis submitted to the Nanyang Technological University in partial fulfillment of the requirement for the degree of Doctor of Philosophy 2014 Abstract Human hair keratins are readily available, easy to extract and eco-friendly materials with promising bioactivities. These properties make them the ideal building blocks for a scaffold. This research was aimed at developing keratin-based materials with an emphasis on keratin hydrogels. We started with keratin extraction from human hair. The optimized extraction method yields 24 ±1% soluble proteins from human hair. The major components were keratins and keratin associated proteins. The most abundant amino acids were cysteine (>10 mol %), Glutamine/ Glutamic acid (13.6 mol %) and Proline (11 mol %). The first application was using CaCl2 induced keratin hydrogels as 2D cell culture substrates. The CaCl2 induced keratin hydrogels were first tested for physical and mechanical properties. In the in vitro experiment, L929 cells seeded on keratin hydrogels had an average proliferation rate 76% compared to cells on collagen hydrogels. -

Damage of Hair Follicle Stem Cells and Alteration of Keratin Expression in External Radiation-Induced Acute Alopecia
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 30: 579-584, 2012 Damage of hair follicle stem cells and alteration of keratin expression in external radiation-induced acute alopecia NAOKI NANASHIMA, KOICHI ITO, TAKASHI ISHIKAWA, MANABU NAKANO and TOSHIYA NAKAMURA Department of Biomedical Sciences, Division of Medical Life Sciences, Hirosaki University Graduate School of Health Sciences, Hirosaki, Japan Received April 4, 2012; Accepted May 28, 2012 DOI: 10.3892/ijmm.2012.1018 Abstract. Alopecia is known as a symptom of acute radia- disturbances and blood and bone marrow disorders are known tion, yet little is known concerning the mechanism of this to occur within several hours to several weeks after 1-6 Gy of phenomenon and the alteration of hair protein profiles. To radiation exposure (4,6). examine this, 6-week-old male C57/BL6 mice were exposed Hair loss is also an effect of ARS, but little is known to 6 Gy of X-ray irradiation, which caused acute alopecia. about the mechanism underlying radiation-induced hair loss. Their hair and skin were collected, and hair proteins were In humans, hair loss is caused by radiation of more than analyzed with liquid chromatography/electrospray-ionization 3 Gy, and almost complete hair loss occurs within weeks of mass spectrometry and immunohistochemistry. No change exposure to 6 Gy (4,6). Since blood stem cells are sensitive to was observed in the composition of major hair keratins, such radiation (7), hair loss is thought to be caused by irradiation- as Krt81, Krt83 and Krt86. However, cytokeratin Krt15 and induced stem cell damage, yet no studies have investigated CD34, which are known as hair follicle stem cell markers, this hypothesis. -

Strand Breaks for P53 Exon 6 and 8 Among Different Time Course of Folate Depletion Or Repletion in the Rectosigmoid Mucosa
SUPPLEMENTAL FIGURE COLON p53 EXONIC STRAND BREAKS DURING FOLATE DEPLETION-REPLETION INTERVENTION Supplemental Figure Legend Strand breaks for p53 exon 6 and 8 among different time course of folate depletion or repletion in the rectosigmoid mucosa. The input of DNA was controlled by GAPDH. The data is shown as ΔCt after normalized to GAPDH. The higher ΔCt the more strand breaks. The P value is shown in the figure. SUPPLEMENT S1 Genes that were significantly UPREGULATED after folate intervention (by unadjusted paired t-test), list is sorted by P value Gene Symbol Nucleotide P VALUE Description OLFM4 NM_006418 0.0000 Homo sapiens differentially expressed in hematopoietic lineages (GW112) mRNA. FMR1NB NM_152578 0.0000 Homo sapiens hypothetical protein FLJ25736 (FLJ25736) mRNA. IFI6 NM_002038 0.0001 Homo sapiens interferon alpha-inducible protein (clone IFI-6-16) (G1P3) transcript variant 1 mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 GALNTL5 NM_145292 0.0001 (GALNT15) mRNA. STIM2 NM_020860 0.0001 Homo sapiens stromal interaction molecule 2 (STIM2) mRNA. ZNF645 NM_152577 0.0002 Homo sapiens hypothetical protein FLJ25735 (FLJ25735) mRNA. ATP12A NM_001676 0.0002 Homo sapiens ATPase H+/K+ transporting nongastric alpha polypeptide (ATP12A) mRNA. U1SNRNPBP NM_007020 0.0003 Homo sapiens U1-snRNP binding protein homolog (U1SNRNPBP) transcript variant 1 mRNA. RNF125 NM_017831 0.0004 Homo sapiens ring finger protein 125 (RNF125) mRNA. FMNL1 NM_005892 0.0004 Homo sapiens formin-like (FMNL) mRNA. ISG15 NM_005101 0.0005 Homo sapiens interferon alpha-inducible protein (clone IFI-15K) (G1P2) mRNA. SLC6A14 NM_007231 0.0005 Homo sapiens solute carrier family 6 (neurotransmitter transporter) member 14 (SLC6A14) mRNA.