Circulating SARS-Cov-2 Variants in Italy, October 2020-March 2021
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
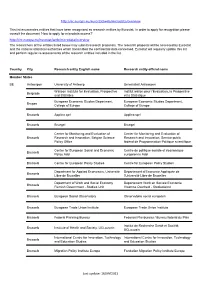
Eurostat: Recognized Research Entity
http://ec.europa.eu/eurostat/web/microdata/overview This list enumerates entities that have been recognised as research entities by Eurostat. In order to apply for recognition please consult the document 'How to apply for microdata access?' http://ec.europa.eu/eurostat/web/microdata/overview The researchers of the entities listed below may submit research proposals. The research proposal will be assessed by Eurostat and the national statistical authorities which transmitted the confidential data concerned. Eurostat will regularly update this list and perform regular re-assessments of the research entities included in the list. Country City Research entity English name Research entity official name Member States BE Antwerpen University of Antwerp Universiteit Antwerpen Walloon Institute for Evaluation, Prospective Institut wallon pour l'Evaluation, la Prospective Belgrade and Statistics et la Statistique European Economic Studies Department, European Economic Studies Department, Bruges College of Europe College of Europe Brussels Applica sprl Applica sprl Brussels Bruegel Bruegel Center for Monitoring and Evaluation of Center for Monitoring and Evaluation of Brussels Research and Innovation, Belgian Science Research and Innovation, Service public Policy Office fédéral de Programmation Politique scientifique Centre for European Social and Economic Centre de politique sociale et économique Brussels Policy Asbl européenne Asbl Brussels Centre for European Policy Studies Centre for European Policy Studies Department for Applied Economics, -

Personal Information Name Francesco Passamonti Birth / Birthplace
Personal Information Name Francesco Passamonti Birth / birthplace October 6, 1966 / Cremona, Italy Citizenship Italian Work address Department of Medicine and Surgery, University of Insubria Hematology, ASST Sette Laghi, Varese Viale Guicciardini - 21100 Varese, Italy Phone +39 (0)332 393 648 Fax +39 (0)332 393 648 Email [email protected] Education 1991 M.D. University of Pavia, School of Medicine, Pavia, Italy 1996 Board in Hematology, University of Pavia, Pavia, Italy Work experience 1991-1998 Physician staff, Division of Hematology, IRCCS Fondazione Policlinico San Matteo, Pavia, Italy 1998-2010 Hematologist, Division of Hematology, IRCCS Fondazione Policlinico San Matteo, Pavia, Italy 1998-2015 Assistant Professor of Hematology, University of Pavia, Pavia, Italy 2010-now Head, Hematology, ASST Sette Laghi, Ospedale di Circolo e Fondazione Macchi, Varese, Italy 2016-2018 Associate Professor of Hematology, University of Insubria, Varese, Italy 2018- Full Professor of Hematology, University of Insubria, Varese, Italy Current clinical activity He leads the Hematology Unit with Stem Cell Transplantation Unit and Trial Unit for research in Hematology. Hematology includes in-patient clinic (12 beds), stem cell transplantation unit (2 beds), Day-Hospital (8 beds), out-patient clinic dedicated to single hematologic malignancies. The clinical activity covers all hematological malignancies and benign disorders. Research interest Biology, diagnosis, prognostication and therapy of myeloid neoplasms, internationally recognized. Principal investigator of several clinical trials in hematology, conducted in accordance to GCP since 1998 Scientific publications Author and co-author of several peer-reviewed scientific articles http://www.pubmed.org in medical journals including New England Journal of Medicine, Lancet Oncology, Blood, Journal of Clinical Oncology, Leukemia, Haematologica, American Journal of Hematology. -

1 Curriculum Vitae of Andrea Vezzulli (March 2018)
Curriculum vitae of Andrea Vezzulli (March 2018) PERSONAL DETAILS: First Name: Andrea Family Name: Vezzulli Gender: Male Place and date of birth: Codogno (LO), Italy. July 19th, 1975 Nationality: Italian Residence address: Via Campagna, 51 26865 San Rocco al Porto (LO) - Italy. Phone: +390377569412(home) +393393541281(mobile) Email: [email protected] EDUCATION: (2006) PhD in Economics, University of Milan, Milan. Thesis title: Bayesian Estimation of Zero Inflated Count Panel Data Models. Methodological Issues and an application to Academic Patenting. Thesis advisor: Prof. Matteo Manera. Examining Committee: Prof. Massimiliano Marcellino, Prof. Eliana La Ferrara, Prof. Alessandra Venturini. (2002) Laurea (MA) in Political Science (major in Economics and Statistics), University of Milan, Milan. Dissertation title: Analysis of a Temporary Work Agency database using Data Mining Techniques. Grade: 106/110. Advisors: Prof. Stefano Maria Iacus, Prof. Giuseppe Porro, Prof. Daniele Checchi. RESEARCH INTERESTS: Banking, innovation and technology transfer, SMEs financing, econometrics. CURRENT POSITION: (December 2016 – present) Assistant Professor (RTDb), Department of Economics, University of Insubria (IT). (April 2013 – present) Research Affiliate (external), ICRIOS – Bocconi University (IT). PREVIOUS POSITIONS: (October 2016 – December 2016) Contract Agent, European Commission Joint Research Centre (JRC), Unit I.1 – Modelling, Indicators and Impact Evaluation - Competence Centre on Composite Indicators and Scoreboards (CC-COIN). (August 2015 – September 2016) Post-Doc Researcher, Department of Economics and Management, University of Pisa (IT). (March 2010 – March 2015) Research Associate (Investigador Auxiliar), UECE/ISEG – University of Lisbon – Lisbon (PT). (March 2009 – March 2010) Post-Doc Research Fellow, Department of Management, University of Bologna, Bologna (IT). (January 2006 – January 2009) Post-Doc Fellow, KITeS/CESPRI – Bocconi University, Milan (IT). -

Esophageal Ph-Impedance Monitoring in Children: Position Paper On
Digestive and Liver Disease 51 (2019) 1522–1536 Contents lists available at ScienceDirect Digestive and Liver Disease journal homepage: www.elsevier.com/locate/dld Position Paper Esophageal pH-impedance monitoring in children: position paper on indications, methodology and interpretation by the SIGENP working group a,b,∗ c,d e f Paolo Quitadamo , Renato Tambucci , Valentina Mancini , Fernanda Cristofori , g h f i Mariella Baldassarre , Licia Pensabene , Ruggiero Francavilla , Giovanni Di Nardo , c j j k b Tamara Caldaro , Paolo Rossi , Saverio Mallardo , Elena Maggiora , Annamaria Staiano , k l m Francesco Cresi , Silvia Salvatore , Osvaldo Borrelli a Department of Pediatrics, A.O.R.N. Santobono-Pausilipon, Naples, Italy b Department of Translational Medical Science,“Federico II”, University of Naples, Italy c Digestive Endoscopy and Surgery Unit, Bambino Gesù Children’s Hospital, IRCCS, Rome, Italy d Department of Biotechnological and Applied Clinical Sciences, University of L’Aquila, L’Aquila, Italy e Department of Pediatrics and Neonatology, Saronno Hospital, Saronno, Italy f Department of Pediatrics, Giovanni XXIII Hospital, Aldo MoroUniversity of Bari, Bari, Italy g Department of Biomedical Science and Human Oncology-neonatology and Nicu section, University “Aldo Moro”, Bari, Italy h Department of Medical and Surgical Sciences, Pediatric Unit, University “Magna Graecia” of Catanzaro, Catanzaro, Italy i NESMOS Department, School of Medicine and Psychology, Sapienza University of Rome, Sant’Andrea University Hospital, Rome, Italy j Department of Pediatrics, Pediatric Gastroenterology and Liver Unit, Sapienza University of Rome, Rome, Italy k Neonatology and Neonatal Intensive Care Unit, Department of Public Health and Pediatrics, University of Turin, Turin, Italy l ¨ Pediatric Department, Ospedale F. -

Curriculum Vitae
CURRICULUM VITAE Surname CONTI, Ph.D., P.D. Faculty of biology and medicine, University of Lausanne Forename Ario Date of birth June 9 th., 1954 Birth place Olivone Nationality Swiss Marital status Married to Roberta Colombi, 1957 2 sons : Nicolò, born 1987 and Matteo, born 1991. Present position Retired since 01 september 2019 Member of the Board of the GRG Gene Technology SA, www.grg-gt.com , Minusio, Switzerland (from 2017) _____________________________________________________________________ Former professional Referee and expert for High Swiss Specialized Schools, Department and academic of Education, Culture and Sports, Bellinzona, Switzerland [2004 – 2019]. positions from 1982 Director, Laboratory Animal Houbandry of the Foundation Cardio- centro Ticino, Lugano – Taverne [2014 – 2020]. Head, Center for Experimental Pathology, Reference Laboratory for Neuroimmunomodulation and supervisor of the Laboratory of histology Istituto Cantonale di Patologia, Centre for Experimental Pathology, P.O.Box, 6601 Locarno 1, Svizzera-Italiana [1982-2004 sept.01] Promoter of the Alpine Institute of Phytopharmacology, Olivone, Switzerland [1997] and Head of the project “Regio Plus” devoted to the development of the Alpine Institute of Phytopharmacology, Congress Center and Farmed Alpine medicinal Plants [1998-2004] Director, Alpine foundation for life sciences – AFLS – Spin off of Educational department, Blenio, State of Ticino, Switzerland (Dipartimento dell’ Educazione, della Cultura e dello Sport, Divisione della Formazione professionale) (Appointed -

Appendix 2 Co-Investigators Name Location Role Contribution Patrizia Accorsi ASST Spedali Civili of Brescia, Italy Site Investi
Appendix 2 Co-investigators Name Location Role Contribution ASST Spedali Civili of Site Participant Patrizia Accorsi Brescia, Italy investigator recruitment/follow-up "V. Buzzi" Children's Site Participant Enrico Alfei Hospital, University of Milan, investigator recruitment/follow-up Italy Meyer Children's University Site Participant Elena Andreucci Hospital, Florence, Italy investigator recruitment/follow-up Fondazione IRCCS Ca' Gianluigi Site Participant Granda, Ospedale Maggiore Ardissino investigator recruitment/follow-up Policlinico, Milan, Italy Oasi Research Institute- Site Participant Emanuela Avola IRCCS, Troina, Italy investigator recruitment/follow-up University of Catania; CNR, Site Participant Rita Barone Catania, Italy investigator recruitment/follow-up Francesco Regional Hospital of Bolzano, Site Participant Benedicenti Italy investigator recruitment/follow-up University Hospital of Ferrara, Site Participant Stefania Bigoni Italy investigator recruitment/follow-up Microcitemie Regional Loredana Site Participant Hospital, Brotzu Hospital, Boccone investigator recruitment/follow-up Cagliari IRCCS Istituto Auxologico Site Participant Maria T. Bonati Italiano, Milan, Italy investigator recruitment/follow-up "V. Buzzi" Children's Site Participant Stefania Bova Hospital, University of Milan, investigator recruitment/follow-up2 Italy Marilena Site Participant University of Messina, Italy Briguglio investigator recruitment/follow-up Site Participant Silvana Briuglia University of Messina, Italy investigator recruitment/follow-up -

Youth Forum 11-12 July, Trieste, ITALY
The following is the list of signatories of the present DECLARATION : 1 Agricultural University of Tirana Albania 2 University of Elbasan Albania 3 Graz University of Technology Austria 4 University of Banja Luka Bosnia and Herzegovina 5 University ‘D zˇemal Bijedi c´’ Mostar Bosnia and Herzegovina 6 University of Mostar Bosnia and Herzegovina 7 University of Split Croatia 8 University of Zadar Croatia 9 Juraj Dobrila University of Pula Croatia 10 Technological Educational Institute of Epirus Greece 11 University of Ioannina Greece 12 Ionian University Greece 13 University of Patras Greece 14 University of Bologna Italy 15 University of Camerino Italy 16 Technical University of Marche Italy TRIESTE 17 University of Trieste Italy 18 University of Udine Italy 19 University of Urbino Italy 20 University of Campania Italy 21 University of Genua Italy 22 University of Foggia Italy DECLARATION 23 University of Insubria Italy 24 University of Modena and Reggio Emilia Italy 25 University of Naples Italy 26 University of Piemonte Orientale Italy 27 University of Teramo Italy 28 University of Palermo Italy 29 University of Milano-Bicocca Italy 30 University of Teramo Italy 31 University of Tuscia Italy 32 University of Venice Ca’Foscari Italy 33 International School for Advanced Studies Italy 34 L’Orientale University of Naples Italy 35 IMT School for Advanced Studies Lucca Italy 36 University of Montenegro Montenegro 37 University of Oradea Romania 38 University Politehnica of Bucharest Romania 39 West University of Timisoara Romania 40 University -

140930 SUPSI Brossura Istituzionale A4 En.Indd
University of Applied Sciences and Arts of Southern Switzerland Coordination University of Applied Sciences and Arts of Southern Switzerland Graphic Design Laboratory of Visual Culture Photographic Credits SUPSI Archive Printing TBS, La Buona Stampa SA, Pregassona This brochure has been printed on FSC certified paper using biological ink, and is therefore eco-friendly © January 2011 Update and reprint: October 2014 SUPSI, Le Gerre, CH-6928 Manno www.supsi.ch © fotopedrazzini.ch University of Applied Sciences and Arts of Southern Switzerland STUDY PROFESSION PASSION Vision SUPSI Profile SUPSI as an agent for change, fostering ◆ Four fields of activity: First-level the development of a region on the University Degree Courses, Continuing cultural cutting edge, embodied by com- Education, Research and Services panies, organisations and professional ◆ The two educational sectors number operators capable of engaging with the more than 10,000 students, from complex structure of socio-economic, Ticino, Switzerland and other countries technological, environmental and cultural phenomena, in accordance with the ◆ 21 Bachelor’s Degree Courses principles of sustainable development. ◆ 14 Master’s Degree Courses Mission ◆ A highly-qualified and skilled teaching body In a society denoted by profound changes, we produce, develop and disseminate ◆ In Ticino, a leading provider in the field knowledge and skills as propelling forces of continuing university education, that are essential for reinforcing the eco- with more than 400 courses nomic, social, technological and artistic ◆ Cutting-edge applied research activities growth of the region, and for contributing to the cultural and ethical development ◆ Services providing business consultancy of both society as a whole and of its indi- and support to the local region vidual members. -

INTERNATIONAL COMPARATIVE ENVIRONMENTAL LAW SEMINAR (Critical Topics in Environmental Law in a Comparative Perspective)
INTERNATIONAL COMPARATIVE ENVIRONMENTAL LAW SEMINAR (Critical Topics in Environmental Law in a Comparative Perspective) COMO, ITALY May 29-June 10, 2016 law.ucdavis.edu/international/study-abroad/como.html CONTENTS The Universities ................................................................................................................................................................................. 2 International Comparative Environmental Law Seminar ................................................................................................. 3 Location—Como, Italy ..................................................................................................................................................................... 3 Curriculum ............................................................................................................................................................................................ 4 Faculty .................................................................................................................................................................................................... 5 Admission, Tuition and Fees ......................................................................................................................................................... 6 Accommodations .............................................................................................................................................................................. 7 Cancellation Policies ....................................................................................................................................................................... -

Mapping Excellence in National Research Systems: the Case of Italy1
MAPPING EXCELLENCE IN NATIONAL RESEARCH SYSTEMS: THE CASE OF ITALY1 GIOVANNI ABRAMO Laboratory for Studies of Research and Technology Transfer, Dept of Management, School of Engineering, University of Rome “Tor Vergata” – Italy and Italian Research Council CIRIACO ANDREA D’ANGELO, FLAVIA DI COSTA Laboratory for Studies of Research and Technology Transfer, Dept of Management, School of Engineering, University of Rome “Tor Vergata” - Italy The study of the concept of “scientific excellence” and the methods for its measurement and evaluation is taking on increasing importance in the development of research policies in many nations. However, scientific excellence results as difficult to define in both conceptual and operational terms, because of its multi-dimensional and highly complex character. The literature on the theme is limited to few studies of an almost pioneering character. This work intends to contribute to the state of the art by exploring a bibliometric methodology which is effective, simple and inexpensive, and which further identifies “excellent” centers of research by beginning from excellence of the individual researchers affiliated with such centers. The study concentrates on the specific case of public research organizations in Italy, analyzing 109 scientific categories of research in the so called “hard” sciences and identifying 157 centers of excellence operating in 60 of these categories. The findings from this first application of the methodology should be considered exploratory and indicative. With a longer period of observation and the addition of further measurements, making the methodology more robust, it can be extended and adapted to a variety of national and supranational contexts, aiding with policy decisions at various levels. -

Società Italiana Di Immunobiologia Comparata E Dello Sviluppo
Società Italiana di Immunobiologia Comparata e dello Sviluppo Italian Association of Developmental and Comparative Immunobiology XXI Incontro Scientifico Aula Granero-Porati, Dipartimento di Biotecnologie e Scienze della Vita (DBSV), via J.H. Dunant 3, Varese PROGRAM 12th February 2020 13.00-15.00 Registration 14.45 Welcome address Session I: Chair- Maria Rosaria Coscia (CNR of Napoli) and Piero G. Giulianini (University of Trieste) Plenary Lecture 15.00 G Scapigliati, A Miccoli, S Picchietti (University of Tuscia) Fish lymphocytes as an equivalent of mammalian innate-type lymphocytes? Communications 15.40 S Picchietti, F Buonocore, A Miccoli, PR Saraceni, AM Fausto, G Scapigliati (University of Tuscia) T lymphocytes in the sea bass (Dicentrarchus labrax) intestine 16.00 M Dara, MG Parisi, C La Corte, D Parrinello P Giulianini, C Manfrin, M Cammarata (University of Palermo) Evolution, adaptation and immune functions of fish F-type lectins. The novelty of FBL from Trematomus bernacchii (Boulenger, 1902) 16.20 A Ametrano, S Greco, U Oreste, M Gerdol, MR Coscia (CNR, Napoli) Investigations on IgT genes of sub-Antarctic fish shed light on the CH2 exon loss 16.40 AM Tolomeo, A Carraro, R Bakiu, S Toppo, M Gerdol, P Irato, D Pellegrino, F Garofalo, P Bisaccia, F Corrà, D Ferro, SP Place, G Santovito (University of Trieste) Too warm or not too warm… Is the antioxidant system of Antarctic fish ready to face climate changes? 17.00 D Rizzotti, S Greco, M Gerdol, C Manfrin, G Santovito, A Pallavicini, PG Giulianini (University of Trieste) Heat stress -

ITALIAN EX LIBRIS USERS ASSOCIATION Annual Report 2013
ITALIAN EX LIBRIS USERS ASSOCIATION Annual Report 2013 Introduction ITALE was established in 1999 to bring together the Italian institutions which had adopted ALEPH. Since January 2007, the Association is open (as in the model provided by IGeLU) to all institutions that use Ex Libris products. Members: 39 (of which 25 are Universities) Membership fee: 250 € (it covers group activities and projects) Products distribution: ALEPH (38, 2 in Saas mode) SFX (21, 1 in SaaS mode) MetaLib (17) MetaLibPlus with Primo (9) DigiTool (1) Primo Local (1), Primo SAaS (8) Bx (2) Since 2009 the local support is provided by the italian subsidiary of Ex Libris (Ex Libris Italy) Activities 2012-2013 After IGeLU Conference in Zurich, a workshop was held in Rome on 15 and 16 November 2012 on the following topic: Choices, plans, guidelines ITALE institutions in times of crisis. That has been an opportunity to talk about: • first experiences with Primo in Italy (in local mode or as SaaS) • implementation experiences of MetaLIB+ • experiences using USTAT • In addition Liliana Morotti, General Manager of Ex Libris Italy, presented the Alma Early Adopters Program for the Italian context. In February 2013, on the occasion of the visit to Italy by Matti Shem Tov, we convened an Assembly of ITALE representatives in Rome: to the President of Ex Libris was presented the current economic situation and the organization of Italian universities. In the years 2009-2012 the Italian universities have dealt with changes in the law (eg new Statutes), organizational and functional changes; they have adopted new evaluation practices and experimented with new ways of negotiation and acquisition of resources at national level (eg.