Exercise and the Skeletal Muscle Epigenome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Roles of Histone Deacetylase 5 and the Histone Methyltransferase Adaptor WDR5 in Myc Oncogenesis
The Roles of Histone Deacetylase 5 and the Histone Methyltransferase Adaptor WDR5 in Myc oncogenesis By Yuting Sun This thesis is submitted in fulfilment of the requirements for the degree of Doctor of Philosophy at the University of New South Wales Children’s Cancer Institute Australia for Medical Research School of Women’s and Children’s Health, Faculty of Medicine University of New South Wales Australia August 2014 PLEASE TYPE THE UNIVERSITY OF NEW SOUTH WALES Thesis/Dissertation Sheet Surname or Family name: Sun First name: Yuting Other name/s: Abbreviation for degree as given in the University calendar: PhD School : School of·Women's and Children's Health Faculty: Faculty of Medicine Title: The Roles of Histone Deacetylase 5 and the Histone Methyltransferase Adaptor WDR5 in Myc oncogenesis. Abstract 350 words maximum: (PLEASE TYPE) N-Myc Induces neuroblastoma by regulating the expression of target genes and proteins, and N-Myc protein is degraded by Fbxw7 and NEDD4 and stabilized by Aurora A. The class lla histone deacetylase HDAC5 suppresses gene transcription, and blocks myoblast and leukaemia cell differentiation. While histone H3 lysine 4 (H3K4) trimethylation at target gene promoters is a pre-requisite for Myc· induced transcriptional activation, WDRS, as a histone H3K4 methyltransferase presenter, is required for H3K4 methylation and transcriptional activation mediated by a histone H3K4 methyltransferase complex. Here, I investigated the roles of HDAC5 and WDR5 in N-Myc overexpressing neuroblastoma. I have found that N-Myc upregulates HDAC5 protein expression, and that HDAC5 represses NEDD4 gene expression, increases Aurora A gene expression and consequently upregulates N-Myc protein expression in neuroblastoma cells. -

Determining HDAC8 Substrate Specificity by Noah Ariel Wolfson A
Determining HDAC8 substrate specificity by Noah Ariel Wolfson A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biological Chemistry) in the University of Michigan 2014 Doctoral Committee: Professor Carol A. Fierke, Chair Professor Robert S. Fuller Professor Anna K. Mapp Associate Professor Patrick J. O’Brien Associate Professor Raymond C. Trievel Dedication My thesis is dedicated to all my family, mentors, and friends who made getting to this point possible. ii Table of Contents Dedication ....................................................................................................................................... ii List of Figures .............................................................................................................................. viii List of Tables .................................................................................................................................. x List of Appendices ......................................................................................................................... xi Abstract ......................................................................................................................................... xii Chapter 1 HDAC8 substrates: Histones and beyond ...................................................................... 1 Overview ..................................................................................................................................... 1 HDAC introduction -

Antigen-Specific Memory CD4 T Cells Coordinated Changes in DNA
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology The Journal of Immunology published online 18 March 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/03/17/jimmun ol.1202267 Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto, Katsumi Ogoshi, Atsushi Sasaki, Jun Abe, Wei Qu, Yoichiro Nakatani, Budrul Ahsan, Kenshiro Oshima, Francis H. W. Shand, Akio Ametani, Yutaka Suzuki, Shuichi Kaneko, Takashi Wada, Masahira Hattori, Sumio Sugano, Shinichi Morishita and Kouji Matsushima J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Author Choice option Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Freely available online through http://www.jimmunol.org/content/suppl/2013/03/18/jimmunol.120226 7.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Author Choice Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. Published March 18, 2013, doi:10.4049/jimmunol.1202267 The Journal of Immunology Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto,*,†,‡ Katsumi Ogoshi,* Atsushi Sasaki,† Jun Abe,* Wei Qu,† Yoichiro Nakatani,† Budrul Ahsan,x Kenshiro Oshima,† Francis H. -

SET-Induced Calcium Signaling and MAPK/ERK Pathway Activation Mediate Dendritic Cell-Like Differentiation of U937 Cells
Leukemia (2005) 19, 1439–1445 & 2005 Nature Publishing Group All rights reserved 0887-6924/05 $30.00 www.nature.com/leu SET-induced calcium signaling and MAPK/ERK pathway activation mediate dendritic cell-like differentiation of U937 cells A Kandilci1 and GC Grosveld1 1Department of Genetics and Tumor Cell Biology, Mail Stop 331, St Jude Children’s Research Hospital, 332 N. Lauderdale, Memphis, TN 38105, USA Human SET, a target of chromosomal translocation in human G1/S transition by allowing cyclin E–CDK2 activity in the leukemia encodes a highly conserved, ubiquitously expressed, presence of p21.11 Second, SET interacts with cyclin B–CDK1.19 nuclear phosphoprotein. SET mediates many functions includ- ing chromatin remodeling, transcription, apoptosis and cell Overexpression of SET inhibits cyclin B-CDK1 activity, which in cycle control. We report that overexpression of SET directs turn, blocks the G2/M transition; this finding suggests a negative 13 differentiation of the human promonocytic cell line U937 along regulatory role for SET in G2/M transition. Overexpression of the dendritic cell (DC) pathway, as cells display typical cell division autoantigen-1 (CDA1), another member of the morphologic changes associated with DC fate and express NAP/SET family, inhibits proliferation and decreases bromo- the DC surface markers CD11b and CD86. Differentiation occurs deoxyuridine uptake in HeLa cells.20 Acidic and basic domains via a calcium-dependent mechanism involving the CaMKII and 20 MAPK/ERK pathways. Similar responses are elicited by inter- of CDA1 show 40% identity and 68% similarity to SET. feron-c (IFN-c) treatment with the distinction that IFN-c signaling We have recently shown that overexpression of SET in the activates the DNA-binding activity of STAT1 whereas SET human promonocytic cell line U937 causes G0/G1 arrest and overexpression does not. -
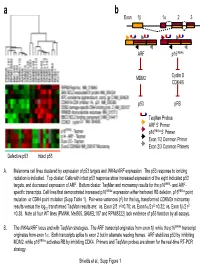
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S. -

The Role of Post-Translational Acetylation and Deacetylation of Signaling Proteins and Transcription Factors After Cerebral Ischemia: Facts and Hypotheses
International Journal of Molecular Sciences Review The Role of Post-Translational Acetylation and Deacetylation of Signaling Proteins and Transcription Factors after Cerebral Ischemia: Facts and Hypotheses Svetlana Demyanenko 1,* and Svetlana Sharifulina 1,2 1 Laboratory of Molecular Neurobiology, Academy of Biology and Biotechnology, Southern Federal University, pr. Stachki 194/1, 344090 Rostov-on-Don, Russia; [email protected] 2 Neuroscience Center HiLife, University of Helsinki, Haartmaninkatu 8, P.O. Box 63, 00014 Helsinki, Finland * Correspondence: [email protected]; Tel.: +7-918-5092185; Fax: +7-863-2230837 Abstract: Histone deacetylase (HDAC) and histone acetyltransferase (HAT) regulate transcription and the most important functions of cells by acetylating/deacetylating histones and non-histone proteins. These proteins are involved in cell survival and death, replication, DNA repair, the cell cycle, and cell responses to stress and aging. HDAC/HAT balance in cells affects gene expression and cell signaling. There are very few studies on the effects of stroke on non-histone protein acetylation/deacetylation in brain cells. HDAC inhibitors have been shown to be effective in protecting the brain from ischemic damage. However, the role of different HDAC isoforms in the survival and death of brain cells after stroke is still controversial. HAT/HDAC activity depends on the acetylation site and the acetylation/deacetylation of the main proteins (c-Myc, E2F1, p53, Citation: Demyanenko, S.; ERK1/2, Akt) considered in this review, that are involved in the regulation of cell fate decisions. Sharifulina, S. The Role of Post-Translational Acetylation and Our review aims to analyze the possible role of the acetylation/deacetylation of transcription factors Deacetylation of Signaling Proteins and signaling proteins involved in the regulation of survival and death in cerebral ischemia. -

Further Investigation of the Potential Anti-Neoplastic, Anti-Inflammatory And
bioRxiv preprint doi: https://doi.org/10.1101/767392; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Further investigation of the potential anti-neoplastic, anti-inflammatory and 2 immunomodulatory actions of phenoxybenzamine using the Broad Institute CLUE 3 platform. 4 5 Mario A. Inchiosa, Jr. 6 Departments of Pharmacology and Anesthesiology, New York Medical College, Valhalla, NY 7 10595 8 9 Short title: Gene expression profiling of phenoxybenzamine in the Broad Institute CLUE 10 platform. 11 12 Keywords: Phenoxybenzamine; gene expression signature predictions; anti-tumor mechanisms; 13 histone deacetylase inhibitors. 14 15 16 17 18 19 1 bioRxiv preprint doi: https://doi.org/10.1101/767392; this version posted September 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 20 21 Abstract: 22 Previous clinical studies with the FDA-approved alpha-adrenergic antagonist, 23 phenoxybenzamine, showed apparent efficacy to reverse the symptoms and disabilities of the 24 neuropathic condition, Complex Regional Pain Syndrome; also, the anatomic spread and 25 intensity of this syndrome has a proliferative character and it was proposed that 26 phenoxybenzamine may have an anti-inflammatory, immunomodulatory mode of action. A 27 previous study gave evidence that phenoxybenzamine had anti-proliferative activity in 28 suppression of growth in several human tumor cell cultures. -

Supplementary Data
Supplemental Material Materials and Methods Immunohistochemistry Primary antibodies used for validation studies include: mouse anti-desmoglein-3 (Cat. # 32-6300, Invitrogen, CA, USA; 1:25), rabbit anti-cytokeratin 4 (Cat. # ab11215, Abcam, Cambridge, MA, USA; 1:100), mouse anti-cytokeratin 16 (Cat. # ab8741, Abcam; 1:25), rabbit anti-desmoplakin antibody (Cat. # ab14418, Abcam; 1:200), mouse anti-vimentin (Cat. # M7020, Dako, Carpinteria, CA, USA; 1:100). Secondary antibodies conjugated with biotin (Vector, Burlingame, CA, USA) were used, diluted to 1:400. Tissues slides containing archival FFPE sections, or tissue micro arrays (TMA) consisting of 508 HNSCC and controls, were dewaxed in SafeClear II (Fisher Scientific, Pittsburgh, PA, USA) hydrated through graded alcohols, immersed in 3% hydrogen peroxide in PBS for 30 min to quench the endogenous peroxidase, and processed for antigen retrieval and immunostaining with the appropriate primary antibodies and biotinylated secondary antibodies as described (1), followed by the avidin-biotin complex method (Vector Stain Elite, ABC kit; Vector). Slides were washed and developed in 3,3'- diaminobenzidine (Sigma FASTDAB tablet; Sigma Chemical) under microscopic control, and counterstained with Mayer's hematoxylin. For each stained TMA the number of positive cells in each core was visually evaluated and the results expressed as a percentage of stained cells/ total number of cells. According to their immunoreactivity the tissues array cores were divided according to tumor differentiation, where the percentage of stained cells in the three tumor classes were scored as more than 5% and less than 25% of the cells stained, 26 to 50%, 51 to 75% or, 75 to 100%. -

Anti-Histone Deacetylase 5 (HDAC5) (PI-16) Antibody Produced in Rabbit
Anti-Histone Deacetylase 5 (HDAC5) (PI-16) Developed in Rabbit, Affinity Isolated Antibody Product Number H 9663 Product Description Class III comprises the yeast Sir2-like proteins. Anti-Histone Deacetylase 5 (HDAC5) (PI-16) is Whereas class I HDACs are ubiquitously expressed, produced in rabbit using as immunogen a synthetic most class II HDACs are tissue-specific.2 The deacet- peptide corresponding to amino acid residues 4-19 of ylase activity of class II HDACs is regulated by sub- HDAC5 with C-terminal added lysine, conjugated to cellular localization.4 The localization of HDAC5 is both KLH. The corresponding sequence is similar in mouse nuclear and cytplasmic. Shuttling to the cytoplasm and human. The antibody is affinity-purified using the occurs during myocyte differentiation; the nuclear immunizing peptide immobilized on agarose. export being stimulated by CaMK phosphorylation at 259 498 Ser and Ser . HDAC5 activity is important for the Anti-Histone Deacetylase 5 (HDAC5) (PI-16) recognizes differentiation of muscle cells by binding, through its N-terminal domain, to the MEF2 protein, thus mouse and human HDAC5. Applications include 6 immunoblotting (~ 124 kDa), immunocytochemistry, and repressing expression of MEF2 down stream genes. immunoprecipitation. Detection of the HDAC5 band by Over expression of HDAC5 in different cancer cells suppresses their growth by induction of apoptosis in a immunoblotting is specifically inhibited with the 7 immunizing peptide. Additional weak bands may be p53-independent manner. detected by immunoblotting in some extract preparations. Reagent The antibody is supplied as a solution in 0.01 M phos- Regulation of gene expression is mediated by several phate buffered saline, pH 7.4, containing 15 mM sodium mechanisms; among them are DNA methylation, ATP- azide. -

Roles for -Synuclein in Gene Expression
G C A T T A C G G C A T genes Review Roles for α-Synuclein in Gene Expression Mahalakshmi Somayaji 1, Zina Lanseur 1, Se Joon Choi 1, David Sulzer 1,2 and Eugene V. Mosharov 1,2,* 1 Departments of Psychiatry and Neurology, Columbia University Medical Center, New York, NY 10032, USA; [email protected] (M.S.); [email protected] (Z.L.); [email protected] (S.J.C.); [email protected] (D.S.) 2 Division of Molecular Therapeutics, New York State Psychiatric Institute, Research Foundation for Mental Hygiene, New York, NY 10032, USA * Correspondence: [email protected] Abstract: α-Synuclein (α-Syn) is a small cytosolic protein associated with a range of cellular com- partments, including synaptic vesicles, the nucleus, mitochondria, endoplasmic reticulum, Golgi apparatus, and lysosomes. In addition to its physiological role in regulating presynaptic function, the protein plays a central role in both sporadic and familial Parkinson’s disease (PD) via a gain-of- function mechanism. Because of this, several recent strategies propose to decrease α-Syn levels in PD patients. While these therapies may offer breakthroughs in PD management, the normal functions of α-Syn and potential side effects of its depletion require careful evaluation. Here, we review recent evidence on physiological and pathological roles of α-Syn in regulating activity-dependent signal transduction and gene expression pathways that play fundamental role in synaptic plasticity. Keywords: Parkinson’s disease; α-Synuclein; gene expression; signal transduction; epigenetics; silencing therapeutics; nuclear receptors; calcium channels Citation: Somayaji, M.; Lanseur, Z.; Choi, S.J.; Sulzer, D.; Mosharov, E.V. -

Hdacs, Histone Deacetylation and Gene Transcription: from Molecular Biology to Cancer Therapeutics
Paola Gallinari et al. npg Cell Research (2007) 17:195-211. npg195 © 2007 IBCB, SIBS, CAS All rights reserved 1001-0602/07 $ 30.00 REVIEW www.nature.com/cr HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics Paola Gallinari1, Stefania Di Marco1, Phillip Jones1, Michele Pallaoro1, Christian Steinkühler1 1Istituto di Ricerche di Biologia Molecolare “P. Angeletti”-IRBM-Merck Research Laboratories Rome, Via Pontina Km 30,600, 00040 Pomezia, Italy Histone deacetylases (HDACs) and histone acetyl transferases (HATs) are two counteracting enzyme families whose enzymatic activity controls the acetylation state of protein lysine residues, notably those contained in the N-terminal extensions of the core histones. Acetylation of histones affects gene expression through its influence on chromatin confor- mation. In addition, several non-histone proteins are regulated in their stability or biological function by the acetylation state of specific lysine residues. HDACs intervene in a multitude of biological processes and are part of a multiprotein family in which each member has its specialized functions. In addition, HDAC activity is tightly controlled through targeted recruitment, protein-protein interactions and post-translational modifications. Control of cell cycle progression, cell survival and differentiation are among the most important roles of these enzymes. Since these processes are affected by malignant transformation, HDAC inhibitors were developed as antineoplastic drugs and are showing encouraging efficacy in cancer patients. Keywords: histone deacetylase, histone, post-translational modification, transcription, histone deacetylase inhibitors, protein acetylation Cell Research (2007) 17:195-211. doi: 10.1038/sj.cr.7310149; published online 27 February 2007 Introduction tone deacetylase (HDAC) family of chromatin-modifying enzymes. -

Supplementary Table 3. Genes Specifically Regulated by Zol (Non-Significant for Fluva)
Supplementary Table 3. Genes specifically regulated by Zol (non-significant for Fluva). log2 Genes Probe Genes Symbol Genes Title Zol100 vs Zol vs Set ID Control (24h) Control (48h) 8065412 CST1 cystatin SN 2,168 1,772 7928308 DDIT4 DNA-damage-inducible transcript 4 2,066 0,349 8154100 VLDLR very low density lipoprotein 1,99 0,413 receptor 8149749 TNFRSF10D tumor necrosis factor receptor 1,973 0,659 superfamily, member 10d, decoy with truncated death domain 8006531 SLFN5 schlafen family member 5 1,692 0,183 8147145 ATP6V0D2 ATPase, H+ transporting, lysosomal 1,689 0,71 38kDa, V0 subunit d2 8013660 ALDOC aldolase C, fructose-bisphosphate 1,649 0,871 8140967 SAMD9 sterile alpha motif domain 1,611 0,66 containing 9 8113709 LOX lysyl oxidase 1,566 0,524 7934278 P4HA1 prolyl 4-hydroxylase, alpha 1,527 0,428 polypeptide I 8027002 GDF15 growth differentiation factor 15 1,415 0,201 7961175 KLRC3 killer cell lectin-like receptor 1,403 1,038 subfamily C, member 3 8081288 TMEM45A transmembrane protein 45A 1,342 0,401 8012126 CLDN7 claudin 7 1,339 0,415 7993588 TMC7 transmembrane channel-like 7 1,318 0,3 8073088 APOBEC3G apolipoprotein B mRNA editing 1,302 0,174 enzyme, catalytic polypeptide-like 3G 8046408 PDK1 pyruvate dehydrogenase kinase, 1,287 0,382 isozyme 1 8161174 GNE glucosamine (UDP-N-acetyl)-2- 1,283 0,562 epimerase/N-acetylmannosamine kinase 7937079 BNIP3 BCL2/adenovirus E1B 19kDa 1,278 0,5 interacting protein 3 8043283 KDM3A lysine (K)-specific demethylase 3A 1,274 0,453 7923991 PLXNA2 plexin A2 1,252 0,481 8163618 TNFSF15 tumor necrosis