Data-Driven Identification of Structural Alerts for Mitigating the Risk of Drug-Induced Human Liver Injuries Ruifeng Liu*, Xueping Yu and Anders Wallqvist*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Carboxylic Acid Derivates
Europaisches Patentamt 0 367 484 J> European Patent Office CO Publication number: A1 Office europeen des brevets © EUROPEAN PATENT APPLICATION A61K 31/29 © Application number: 89310994.2 © int. ci.5: C07D 233/64 , C07D 277/42 C07D 277/28 @ Date of filing: 25.10.89 C07D 295/08 C07D 249/14 C07D 211/20 C07D 417/04 © Priority: 26.10.88 GB 8825058 © Applicant: GLAXO GROUP LIMITED 26.06.89 GB 8914631 Clarges House 6-12 Clarges Street London W1Y8DH(GB) © Date of publication of application: 09.05.90 Bulletin 90/19 © Inventor: Clitherow, John Watson 54, Gilders © Designated Contracting States: Sawbridgeworth Hertfordshire(GB) AT BE CH DE ES FR GB GR IT LI LU NL SE © Representative: Marchant, James Ian et al Elkington and Fife Beacon House 113 Kingsway London WC2B 6PP(GB) © Carboxylic acid derivates. © The invention relates to salts formed between basic H2-receptor antagonists and a complex of bismuth with a carboxylic acid, and solvate of such salts, excluding salts in which the basic H2-receptor antagonist is ranitidine. Examples of suitable carboxylic acids are citric acid and tartaric acid. Examples of basic rVreceptor antagonists are cimetidine, sufotidine famotidine and nizatidine. The salts ares useful in the treatment of gastrointestinal disorders, particularly gastroduodenal conditions. The salts show the antisecretory activity associated with the basic H2-receptor antagonist together with antibacterial activity against Campylobacter pylori and they also possess cytoprotective properties. 00 (0 a. LU Xerox Copy Centre EP 0 367 484 A1 CARBOXYLIC ACID DERIVATIVES This invention relates to salts of compounds having antagonist activity at histamine H2- receptors, to a process for the preparation thereof, to pharmaceutical compositions containing them and to their use in therapeutics. -

Pharmaceutical Appendix to the Tariff Schedule 2
Harmonized Tariff Schedule of the United States (2007) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2007) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. ABACAVIR 136470-78-5 ACIDUM LIDADRONICUM 63132-38-7 ABAFUNGIN 129639-79-8 ACIDUM SALCAPROZICUM 183990-46-7 ABAMECTIN 65195-55-3 ACIDUM SALCLOBUZICUM 387825-03-8 ABANOQUIL 90402-40-7 ACIFRAN 72420-38-3 ABAPERIDONUM 183849-43-6 ACIPIMOX 51037-30-0 ABARELIX 183552-38-7 ACITAZANOLAST 114607-46-4 ABATACEPTUM 332348-12-6 ACITEMATE 101197-99-3 ABCIXIMAB 143653-53-6 ACITRETIN 55079-83-9 ABECARNIL 111841-85-1 ACIVICIN 42228-92-2 ABETIMUSUM 167362-48-3 ACLANTATE 39633-62-0 ABIRATERONE 154229-19-3 ACLARUBICIN 57576-44-0 ABITESARTAN 137882-98-5 ACLATONIUM NAPADISILATE 55077-30-0 ABLUKAST 96566-25-5 ACODAZOLE 79152-85-5 ABRINEURINUM 178535-93-8 ACOLBIFENUM 182167-02-8 ABUNIDAZOLE 91017-58-2 ACONIAZIDE 13410-86-1 ACADESINE 2627-69-2 ACOTIAMIDUM 185106-16-5 ACAMPROSATE 77337-76-9 -

Cytochrome P450s and Other Enzymes in Drug Metabolism and Toxicity Submitted: December 15 , 2005 ; Accepted: January 12 , 2006 ; Published: March 10, 2006 F
The AAPS Journal 2006; 8 (1) Article 12 (http://www.aapsj.org). Themed Issue: NIDA/AAPS Symposium on Drugs of Abuse: Mechanisms of Toxicity, Toxicokinetics and Medical Consequences, November 4-5, 2005 Guest Editor - Rao S. Rapaka Cytochrome P450s and Other Enzymes in Drug Metabolism and Toxicity Submitted: December 15 , 2005 ; Accepted: January 12 , 2006 ; Published: March 10, 2006 F. Peter Guengerich1 1 Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, TN 37232 A BSTRACT C HALLENGE OF METABOLISM IN DRUG The cytochrome P450 (P450) enzymes are the major cata- DEVELOPMENT lysts involved in the metabolism of drugs. Bioavailability The numbers related to success in producing new medicines and toxicity are 2 of the most common barriers in drug devel- are very challenging. Even with some discrimination in the opment today, and P450 and the conjugation enzymes can selection of chemicals for leads, ~104 compounds are tested infl uence these effects. The toxicity of drugs can be con- to produce one compound that reaches the market. The aver- sidered in 5 contexts: on-target toxicity, hypersensitivity age cost of taking a drug to the market stage is $800 million and immunological reactions, off-target pharmacology, bio- to $1 billion, and only 1 in 3 drugs that does reach the mar- activation to reactive intermediates, and idiosyncratic drug ket is profi table. Compounding the problems are social and reactions. The chemistry of bioactivation is reasonably well political pressures to both reduce the costs of drugs and pro- understood, but the mechanisms underlying biological re - vide nearly absolute safety. -
Effervescent H2 Blocker Formulation
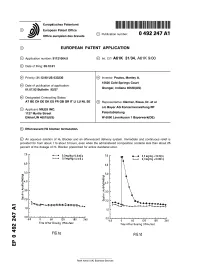
Europaisches Patentamt European Patent Office © Publication number: 0 492 247 A1 Office europeen des brevets EUROPEAN PATENT APPLICATION © Application number: 91121064.9 © int. CIA A61K 31/34, A61K 9/00 @ Date of filing: 09.12.91 ® Priority: 21.12.90 US 633230 @ Inventor: Paulos, Manley A. 15626 Cold Springs Court @ Date of publication of application: Indiana 01.07.92 Bulletin 92/27 Granger, 46530(US) © Designated Contracting States: AT BE CH DE DK ES FR GB GR IT LI LU NL SE © Representative: Danner, Klaus, Dr. et al c/o Bayer AG Konzernverwaltung RP © Applicant: MILES INC. 1127 Myrtle Street Patentabteilung ElkhartJN 4651 5(US) W-5090 Leverkusen 1 Bayerwerk(DE) (§v Effervescent H2 blocker formulation. © An aqueous solution of H2 Blocker and an effervescent delivery system. Immediate and continuous relief is provided for from about 1 to about 3 hours, even when the administered composition contains less than about 25 percent of the dosage of H2 Blocker prescribed for active duodenal ulcer. 7.0 r 0,3mg/kg+0,0mEq 7.0 0,3 mg/kg + 32mEq 0,0mg/kg+0,0mEq 0,0mg/kg +0,0mE q 6.0 6,0 5,0 5,0 Xa. *4.0 :4.0 ^3.0 :3,0 1^2,0 h is 2,0 1.0 h 1.0 ■ 0.0 _i i ■ ■ 0.0 CM -60 0 60 120 180 240 -60 0 60 120 180 240 Time After Dosing (Minutes) CM Time After Dosing (Minutes) Oi FIG.1Q FIG.Id Rank Xerox (UK) Business Services EP 0 492 247 A1 Background of the Invention Histamine H2 receptor blocking agents (referred to herein as H2 Blockers) are a class of drugs which act as antagonists of the histamine H2 receptor. -

Ranitidine Protocol – Final Confidential Version 1.0 – 17Th January 2020 PASS Information
RANITIDINE AND OTHER HISTAMINE-H2-RECEPTOR ANTAGONISTS – A DRUG UTILISATION STUDY Principal Investigators Katia Verhamme, MD, PhD Department of Medical Informatics EMC – Rotterdam Peter Rijnbeek, PhD Department of Medical Informatics EMC - Rotterdam Document Status Final Version 1.0 Date of final version of the study 17th January 2020 report EU PAS register number 1 EMA TENDER – Ranitidine protocol – Final Confidential Version 1.0 – 17th January 2020 PASS information Title Ranitidine and other histamine H2-receptor antagonists – a drug utilisation study Protocol version identifier V1.0 Final Date of last version of protocol 2020-01-17 EU PAS register number Active Ingredient Ranitidine Cimetidine Famotidine Nizatidine Niperotidine Roxatidine Ranitidine bismuth citrate Lafutidine Medicinal product Ranitidine Product reference Procedure number Marketing authorisation holder(s) Joint PASS Research question and objectives Ranitidine is a competitive and reversible inhibitor of the action of histamine and indicated for the management of peptic ulceration, Gastro-Esophageal Reflux Disease (GERD), reflux oesophagitis and Zollinger-Ellison syndrome. Results of a preliminary laboratory analysis have shown the presence of N-Nitrosodimethylamine (NDMA), a human carcinogen, in ranitidine. With this DUS, we aim to determine drug utilisation and prescription patterns of medicinal products containing H2-receptor antagonists. These data will give insight on the number of patients using ranitidine and thus potentially at risk of NDMA. In particular we will: 1. Study the prevalence and incidence prescribing of H2-receptor antagonists as a class and by individual drug 2 EMA TENDER – Ranitidine protocol – Final Confidential Version 1.0 – 17th January 2020 2. Explore the characteristics of H2-receptor antagonist use with regard to age (10-years age categories), sex, formulation, daily dose, duration and cumulative exposure by class level and individual ingredient 3. -

Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DIX to the HTSUS—Continued
20558 Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DEPARMENT OF THE TREASURY Services, U.S. Customs Service, 1301 TABLE 1.ÐPHARMACEUTICAL APPEN- Constitution Avenue NW, Washington, DIX TO THE HTSUSÐContinued Customs Service D.C. 20229 at (202) 927±1060. CAS No. Pharmaceutical [T.D. 95±33] Dated: April 14, 1995. 52±78±8 ..................... NORETHANDROLONE. A. W. Tennant, 52±86±8 ..................... HALOPERIDOL. Pharmaceutical Tables 1 and 3 of the Director, Office of Laboratories and Scientific 52±88±0 ..................... ATROPINE METHONITRATE. HTSUS 52±90±4 ..................... CYSTEINE. Services. 53±03±2 ..................... PREDNISONE. 53±06±5 ..................... CORTISONE. AGENCY: Customs Service, Department TABLE 1.ÐPHARMACEUTICAL 53±10±1 ..................... HYDROXYDIONE SODIUM SUCCI- of the Treasury. NATE. APPENDIX TO THE HTSUS 53±16±7 ..................... ESTRONE. ACTION: Listing of the products found in 53±18±9 ..................... BIETASERPINE. Table 1 and Table 3 of the CAS No. Pharmaceutical 53±19±0 ..................... MITOTANE. 53±31±6 ..................... MEDIBAZINE. Pharmaceutical Appendix to the N/A ............................. ACTAGARDIN. 53±33±8 ..................... PARAMETHASONE. Harmonized Tariff Schedule of the N/A ............................. ARDACIN. 53±34±9 ..................... FLUPREDNISOLONE. N/A ............................. BICIROMAB. 53±39±4 ..................... OXANDROLONE. United States of America in Chemical N/A ............................. CELUCLORAL. 53±43±0 -

2 12/ 35 74Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date 22 March 2012 (22.03.2012) 2 12/ 35 74 Al (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A61K 9/16 (2006.01) A61K 9/51 (2006.01) kind of national protection available): AE, AG, AL, AM, A61K 9/14 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, (21) International Application Number: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/EP201 1/065959 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, 14 September 201 1 (14.09.201 1) ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, (25) Filing Language: English RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, (26) Publication Language: English TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 61/382,653 14 September 2010 (14.09.2010) US (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (71) Applicant (for all designated States except US): GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, NANOLOGICA AB [SE/SE]; P.O Box 8182, S-104 20 ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, Stockholm (SE). -

Niperotidine Hydrochloride
Metopimazine/Nizatidine 1751 ataxia, confusion, disorientation, dizziness, euphoria, dysphoria, little effect on the metabolism of other drugs. However, hallucinations, psychosis, depression, headache, decreased con- like other H2-antagonists its effects on gastric pH may centration, blurred vision, sleep disturbances, decreased coordi- O affect the absorption of some other drugs. nation, and tremors. Adverse cardiovascular reactions including -O N+ hypotension, orthostatic hypotension, and tachycardia have oc- curred. Gastrointestinal disturbances, decreased appetite, and ab- HN Pharmacokinetics dominal pain have also been reported. Nizatidine is readily and almost completely absorbed O HN Precautions S from the gastrointestinal tract. The bioavailability of Nabilone is extensively metabolised and largely excreted in bile, O CH3 nizatidine after oral doses exceeds 70% and may be and therefore is not recommended in patients with severe hepatic O N slightly increased by the presence of food. It is widely impairment. It should be used with caution in patients with a his- CH3 distributed and is about 35% bound to plasma proteins. tory of psychiatric disorders or depression, or those with hyper- tension or heart disease. (niperotidine) The elimination half-life of nizatidine is 1 to 2 hours Because of the possibility of CNS depression, patients should be and is prolonged in renal impairment. Nizatidine is warned not to drive or operate machinery. Profile partly metabolised in the liver: nizatidine N-2-oxide, Niperotidine hydrochloride is a histamine H2-receptor antagonist nizatidine S-oxide, and N-2-monodesmethylnizatidine The possibility of dependence similar to that of cannabis should with general properties similar to those of cimetidine (p.1716). be borne in mind. -

Histamine Receptor
Histamine Receptor Histamine Receptors are a class of G protein-coupled receptors with histamine as their endogenous ligand. There are four known histamine receptors: H1 receptor, H2 receptor, H3 receptor, H4 receptor. The H1 receptor is a histamine receptor belonging to the family of Rhodopsin-like G-protein-coupled receptors. This receptor, which is activated by the biogenic amine histamine, is expressed throughout the body, to be specific, in smooth muscles, on vascular endothelial cells, in the heart, and in the central nervous system. H2 receptors are positively coupled to adenylate cyclase via Gs. It is a potent stimulant of cAMP production, which leads to activation of Protein Kinase A. Histamine H3 receptors are expressed in the central nervous system and to a lesser extent the peripheral nervous system, where they act asautoreceptors in presynaptic histaminergic neurons, and also control histamine turnover by feedback inhibition of histamine synthesis and release. The Histamine H4 receptor has been shown to be involved in mediating eosinophil shape change and mast cell chemotaxis. www.MedChemExpress.com 1 Histamine Receptor Inhibitors & Modulators (±)-Methotrimeprazine (D6) (±)-Tazifylline (dl-Methotrimeprazine D6) Cat. No.: HY-19489S Cat. No.: HY-U00018 Bioactivity: (±)-Methotrimeprazine (D6) is the deuterium labeled Bioactivity: (±)-Tazifylline is a potent, selective and long-acting Methotrimeprazine, which is a D3 dopamine and Histamine H1 histamine H1 receptor antagonist. receptor antagonist. Purity: >98% Purity: >98% Clinical Data: No Development Reported Clinical Data: No Development Reported Size: 1 mg Size: 1 mg, 5 mg, 10 mg, 20 mg ABT-239 Acrivastine Cat. No.: HY-12195 (BW825C) Cat. No.: HY-B1510 Bioactivity: ABT-239 is a novel, highly efficacious, Bioactivity: Acrivastine (BW825C) is a short acting histamine 1 non-imidazole class of H3R antagonist and a transient receptor antagonist for the treatment of allergic rhinitis. -

Stembook 2018.Pdf
The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 WHO/EMP/RHT/TSN/2018.1 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances. Geneva: World Health Organization; 2018 (WHO/EMP/RHT/TSN/2018.1). Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -

High Daily Dose and Being a Substrate of Cytochrome P450 Enzymes Are Two Important Predictors of Drug-Induced Liver Injury
High daily dose and being a substrate of Cytochrome P450 enzymes are two important predictors of drug-induced liver injury Ke Yu, Xingchao Geng, Minjun Chen, Jie Zhang, Bingshun Wang, Katarina Ilic and Weida Tong Journal title: Drug metabolism and disposition SUPPLEMENTARY TABLE 1 Drugs-substrates, inhibitors, and inducers of CYP enzymes with the daily dose and lipophilicity SUPPLEMENTARY TABLE 1 Drugs-substrates, inhibitors, and inducers of CYP enzymes with the daily dose and lipophilicity 128 Most-DILI- Daily dose CYP substrates* CYP inducers CYP inhibitors LogP concern drugs (mg) Abacavir DNF DNF DNF 0.8 600 Acarbose DNF 2E1 DNF -6.8 300 Acitretin DNF 26A1 DNF 5.7 35 Alaproclate DNF DNF DNF 2.8 200 Unspecified CYP Alclofenac DNF DNF 3000 isoforms 2.8 Alpidem 3A4 DNF DNF 5.5 150 Ambrisentan 3A4, 3A5, 2C19 DNF DNF 3.7 5 Unspecified CYP Amineptine DNF DNF 150 isoforms 1.1 Aminosalicylic DNF DNF 11B2 12000 Acid 0.5 1A1, 1A2, 2C8, 1A2, 2C9, 2D6, 3A4, Amiodarone DNF 200 2C19, 2D6, 3A4 3A5, 3A7 7.2 Aplaviroc 3A4,2C19 DNF DNF 3.1 200 Bendazac DNF DNF DNF 3.3 300 Unspecified CYP Benoxaprofen 1A1, 1A2 DNF 300 isoforms 4.1 Benzarone DNF DNF DNF 4.1 400 Benzbromarone 2C9 DNF 2C9 5.6 100 Benziodarone DNF DNF 2D6 5.3 600 Bicalutamide 3A4 DNF 2C9, 2C19, 2D6, 3A4 50 2.9 Bosentan 2C9, 3A4 2C9, 3A4 DNF 3.9 125 Unspecified CYP Bromfenac DNF DNF 150 isoforms 2.9 Busulfan 3A4 DNF DNF -0.3 240 2B6, 2C9, 2C19, Carbamazepine 1A2, 2C8, 3A4, 3A7 1A2, 2C19 1000 3A4 2.7 Carbidopa DNF DNF DNF 0.4 100 Chlormezanone DNF DNF DNF 1.6 600 1A1, 1A2, 2A6, 2D6, Chlorzoxazone -

Harmonized Tariff Schedule of the United States (2004) -- Supplement 1 Annotated for Statistical Reporting Purposes
Harmonized Tariff Schedule of the United States (2004) -- Supplement 1 Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2004) -- Supplement 1 Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. Product CAS No. Product CAS No. ABACAVIR 136470-78-5 ACEXAMIC ACID 57-08-9 ABAFUNGIN 129639-79-8 ACICLOVIR 59277-89-3 ABAMECTIN 65195-55-3 ACIFRAN 72420-38-3 ABANOQUIL 90402-40-7 ACIPIMOX 51037-30-0 ABARELIX 183552-38-7 ACITAZANOLAST 114607-46-4 ABCIXIMAB 143653-53-6 ACITEMATE 101197-99-3 ABECARNIL 111841-85-1 ACITRETIN 55079-83-9 ABIRATERONE 154229-19-3 ACIVICIN 42228-92-2 ABITESARTAN 137882-98-5 ACLANTATE 39633-62-0 ABLUKAST 96566-25-5 ACLARUBICIN 57576-44-0 ABUNIDAZOLE 91017-58-2 ACLATONIUM NAPADISILATE 55077-30-0 ACADESINE 2627-69-2 ACODAZOLE 79152-85-5 ACAMPROSATE 77337-76-9 ACONIAZIDE 13410-86-1 ACAPRAZINE 55485-20-6 ACOXATRINE 748-44-7 ACARBOSE 56180-94-0 ACREOZAST 123548-56-1 ACEBROCHOL 514-50-1 ACRIDOREX 47487-22-9 ACEBURIC ACID 26976-72-7