PBX Mesenchyme Supplemental Figs 12132017
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes S
Supplemental material to this article can be found at: http://molpharm.aspetjournals.org/content/suppl/2017/03/02/mol.116.107904.DC1 1521-0111/91/5/518–532$25.00 https://doi.org/10.1124/mol.116.107904 MOLECULAR PHARMACOLOGY Mol Pharmacol 91:518–532, May 2017 Copyright ª 2017 by The American Society for Pharmacology and Experimental Therapeutics The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes s Katharina Simon, Nicole Merten, Ralf Schröder, Stephanie Hennen, Philip Preis, Nina-Katharina Schmitt, Lucas Peters, Ramona Schrage,1 Celine Vermeiren, Michel Gillard, Klaus Mohr, Jesus Gomeza, and Evi Kostenis Molecular, Cellular and Pharmacobiology Section, Institute of Pharmaceutical Biology (K.S., N.M., Ral.S., S.H., P.P., N.-K.S, L.P., J.G., E.K.), Research Training Group 1873 (K.S., E.K.), Pharmacology and Toxicology Section, Institute of Pharmacy (Ram.S., K.M.), University of Bonn, Bonn, Germany; UCB Pharma, CNS Research, Braine l’Alleud, Belgium (C.V., M.G.). Downloaded from Received December 16, 2016; accepted March 1, 2017 ABSTRACT Pairing orphan G protein–coupled receptors (GPCRs) with their using eight distinct functional assay platforms based on label- cognate endogenous ligands is expected to have a major im- free pathway-unbiased biosensor technologies, as well as molpharm.aspetjournals.org pact on our understanding of GPCR biology. It follows that the canonical second-messenger or biochemical assays. Appraisal reproducibility of orphan receptor ligand pairs should be of of GPR17 activity can be accomplished with neither the coapplica- fundamental importance to guide meaningful investigations into tion of both ligand classes nor the exogenous transfection of partner the pharmacology and function of individual receptors. -

Regulation of Procollagen Amino-Propeptide Processing During Mouse Embryogenesis by Specialization of Homologous ADAMTS Protease
DEVELOPMENT AND DISEASE RESEARCH ARTICLE 1587 Development 133, 1587-1596 (2006) doi:10.1242/dev.02308 Regulation of procollagen amino-propeptide processing during mouse embryogenesis by specialization of homologous ADAMTS proteases: insights on collagen biosynthesis and dermatosparaxis Carine Le Goff1, Robert P. T. Somerville1, Frederic Kesteloot2, Kimerly Powell1, David E. Birk3, Alain C. Colige2 and Suneel S. Apte1,* Mutations in ADAMTS2, a procollagen amino-propeptidase, cause severe skin fragility, designated as dermatosparaxis in animals, and a subtype of the Ehlers-Danlos syndrome (dermatosparactic type or VIIC) in humans. Not all collagen-rich tissues are affected to the same degree, which suggests compensation by the ADAMTS2 homologs ADAMTS3 and ADAMTS14. In situ hybridization of Adamts2, Adamts3 and Adamts14, and of the genes encoding the major fibrillar collagens, Col1a1, Col2a1 and Col3a1, during mouse embryogenesis, demonstrated distinct tissue-specific, overlapping expression patterns of the protease and substrate genes. Adamts3, but not Adamts2 or Adamts14, was co-expressed with Col2a1 in cartilage throughout development, and with Col1a1 in bone and musculotendinous tissues. ADAMTS3 induced procollagen I processing in dermatosparactic fibroblasts, suggesting a role in procollagen I processing during musculoskeletal development. Adamts2, but not Adamts3 or Adamts14, was co-expressed with Col3a1 in many tissues including the lungs and aorta, and Adamts2–/– mice showed widespread defects in procollagen III processing. Adamts2–/– mice had abnormal lungs, characterized by a decreased parenchymal density. However, the aorta and collagen fibrils in the aortic wall appeared normal. Although Adamts14 lacked developmental tissue-specific expression, it was co-expressed with Adamts2 in mature dermis, which possibly explains the presence of some processed skin procollagen in dermatosparaxis. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

Cloning of ADAMTS2 Gene and Colony Formation Effect of ADAMTS2 in Saos-2 Cell Line Under Normal and Hypoxic Conditions, ADYU J SCI, 10(2), 413-426
Aydogan Türkoğlu & Gültekin Tosun (2020) Cloning of ADAMTS2 Gene and Colony Formation Effect of ADAMTS2 in Saos-2 Cell Line Under Normal and Hypoxic Conditions, ADYU J SCI, 10(2), 413-426 Cloning of ADAMTS2 Gene and Colony Formation Effect of ADAMTS2 in Saos-2 Cell Line Under Normal and Hypoxic Conditions Sümeyye AYDOGAN TÜRKOĞLU1,*, Sinem GÜLTEKİN TOSUN2 1Balıkesir University, Faculty of Science and Literature, Department of Molecular Biology and Genetics, Balıkesir, Turkey [email protected], ORCID: 0000-0003-1754-0700 2Erciyes University, Institute of Health Sciences, Faculty of Veterinary Medicine, Department of Genetics, Kayseri, Turkey [email protected], ORCID: 0000-0002-3927-0089 Received: 03.05.2020 Accepted: 25.09.2020 Published: 30.12.2020 Abstract ADAMTS2 (a disintegrin and metalloproteinase with thrombospondin motifs 2), an N- propeptidase isoenzyme, is an enzyme involved in collagen biosynthesis by providing the amino ends of procollagen to be cut away. ADAMTS2 has anti-angiogenic activity as well as provides the processing of collagen. With this activity, it has become a target in cancer studies. Hypoxic regulation is a process that affects the expression of a large number of genes at the cellular level. Within the scope of our study, the cloning of the ADAMTS2 gene and its expression in Saos-2 (human bone carcinoma) cell line were performed ectopically. For this purpose, the transient transfection of the expression vector containing ADAMTS2 coding sequence was transfected by the calcium-phosphate precipitation method. Recombinant ADAMTS2 mRNA expression was checked by Real-Time PCR in Saos-2 cells. It was observed that there was a 50-fold increase in ADAMTS2 mRNA expression in the transfected Saos-2 cells compared to the control group. -
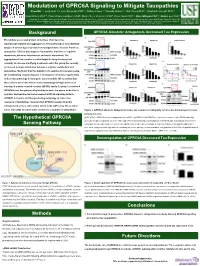
Background the Hypothetical GPRC6A Sensing Pathway
Modulation of GPRC6A Signaling to Mitigate Tauopathies Chao Ma1,2, Jerry Hunt1,3, Leslie Sandusky PhD1,3, William Fraser1,3, Ronald Alvarez1,3, Dali Zheng PhD1,4, Siddharth Kamath PhD1,2, Chad Dickey PhD1,4, Hans Bräuner-Osborne PhD5, Daniel Sejer Pedersen PhD5, Kevin Nash PhD1,2, Dave Morgan PhD1,2, Daniel Lee PhD1,3 1:Byrd Alzheimer‘s Institute, University of South Florida, Tampa, FL 33613, USA. 2:Morsani College of Medicine, Department of Molecular Pharmacology and Physiology, University of South Florida, Tampa, FL 33612, USA. 3:College of Pharmacy, Department of Pharmaceutical Sciences, University of South Florida, Tampa, FL 33612, USA. 4:Morsani College of Medicine, Department of Molecular Medicine, University of South Florida, Tampa, FL 33612, USA. 5:Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Jagtvej 162, 2100 Copenhagen, Denmark Background GPRC6A Allosteric Antagonists Decreased Tau Expression Microtubule associated protein named tau, often becomes hyperphosphorylated and aggregates to form pathological neurofibrillary tangles in several age-associated neurodegenerative diseases known as tauopathies. Clinical phenotypes of tauopathies manifest as cognitive impairment, behavior disturbances and motor impairment. The aggregation of tau remains a central target for drug discovery, but currently no disease-modifying treatments exist. Our group has recently uncovered a unique interaction between L-arginine metabolism and tauopathies. We found that the depletion of L-arginine by overexpressing the metabolizing enzyme arginase 1 and arginine deiminase significantly reduced tau pathology in transgenic mouse models. We speculate that these effects associate with increased autophagy through amino acid sensing. G protein-coupled receptor (GPCR), family C, group 6, member A (GPRC6A) was deorphanized by binding to basic L-α amino acids, like L- arginine. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Novel Types of Mutation Responsible for the Dermatosparactic Type of Ehlers–Danlos Syndrome (Type VIIC) and Common Polymorphisms in the ADAMTS2 Gene
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Novel Types of Mutation Responsible for the Dermatosparactic Type of Ehlers–Danlos Syndrome (Type VIIC) and Common Polymorphisms in the ADAMTS2 Gene Alain Colige,à Lieve Nuytinck,w Ingrid Hausser,z Anthonie J. van Essen,y Marc Thiry,z Christian Herens,# Lesley C. Ade` s,Ãà Fransiska Malfait,w Anne De Paepe,w Peter Franck,ww Gerhard Wolff,zz JanC.Oosterwijk,y J. H. Sillevis Smitt,yy Charles M. Lapie` re,à and Betty V. Nusgensà ÃLaboratory of Connective Tissues Biology, GIGA Research Center, University of Lie` ge, Lie` ge, Belgium; wCentrum voor Medische Genetica, Universitair Ziekenhuis, University of Gent, Gent, Belgium; zElectron Microscopic Laboratory, Department of Dermatology, University Heidelberg, Heidelberg, Germany; yDepartment of Clinical Genetics, University Medical Center, Groningen, The Netherlands; zLaboratoire de Biologie cellulaire et tissulaire, University of Lie` ge, Lie` ge, Belgium; #Center for Human Genetics, University of Lie` ge, Lie` ge, Belgium; ÃÃDepartment of Clinical Genetics, The Children’s Hospital at Westmead, and Discipline of Paediatrics and Child Health, University of Sydney, Sydney, Australia; wwDepartment of Pediatrics, University Freiburg, Freiburg, Germany; zzInstitute of Human Genetics and Anthropology, University Freiburg, Freiburg, Germany; yyDepartment of Dermatology, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands Ehlers–Danlos syndrome (EDS) type VIIC, or dermatosparactic type, is a recessively inherited connective tissue disorder characterized, among other symptoms, by an extreme skin fragility resulting from mutations inactivating ADAMTS-2, an enzyme excising the aminopropeptide of procollagens type I, II, and III. -

An Investigation of ADAM-Like Decysin 1 in Macrophage-Mediated Inflammation and Crohn’S Disease
An Investigation of ADAM-like Decysin 1 in Macrophage-mediated Inflammation and Crohn’s Disease By Nuala Roisin O’Shea A thesis submitted to UCL for the degree of Doctor of Philosophy Division of Medicine Declaration I, Nuala Roisin O’Shea, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. 2 Abstract Crohn’s disease (CD) is now recognised as a defective host response to bacteria in genetically susceptible individuals. The role of innate immunity and impaired bacterial clearance are widely accepted. In this thesis the role of ADAM-like, Decysin-1 (ADAMDEC1) in macrophage-mediated inflammation and gut mucosal immunity is explored. Using transcriptomic analysis of monocyte derived macrophages (MDM) ADAMDEC1 was identified as grossly under expressed in a subset of patients with CD. ADAMDEC1 was found to be highly selective to the intestine, peripheral blood monocyte-derived and lamina propria macrophages. It was shown to be an inflammatory response gene, upregulated in response to bacterial antigens and inflammation. ADAMDEC1 was expressed in prenatal and germ free mice, demonstrating exposure to a bacterial antigen is not a prerequisite for expression. Adamdec1 knock out mice were used to investigate the role of ADAMDEC1 in vivo. Adamdec1-/- mice displayed an increased susceptibility to dextran sodium sulphate (DSS), Citrobacter rodentium and Salmonella typhimurium induced colitis. In Adamdec1-/- mice, bacterial translocation and systemic infection were increased in bacterial models of colitis. These results suggest that individuals with grossly attenuated expression of ADAMDEC1 may be at an increased risk of developing intestinal inflammation as a consequence of an impaired ability to handle enteric bacterial pathogens. -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

Proteomic Analysis of Human Osteoarthritis Synovial Fluid
Balakrishnan et al. Clinical Proteomics 2014, 11:6 http://www.clinicalproteomicsjournal.com/content/11/1/6 CLINICAL PROTEOMICS RESEARCH Open Access Proteomic analysis of human osteoarthritis synovial fluid Lavanya Balakrishnan1,2, Raja Sekhar Nirujogi1,3, Sartaj Ahmad1,4, Mitali Bhattacharjee1,5, Srikanth S Manda1,3, Santosh Renuse1,5, Dhanashree S Kelkar1,5, Yashwanth Subbannayya1,6, Rajesh Raju1, Renu Goel1,2, Joji Kurian Thomas1,5, Navjyot Kaur7, Mukesh Dhillon7, Shantal Gupta Tankala7, Ramesh Jois8, Vivek Vasdev9, YL Ramachandra2, Nandini A Sahasrabuddhe1, TS Keshava Prasad1,3,4, Sujatha Mohan10, Harsha Gowda1, Subramanian Shankar7* and Akhilesh Pandey11,12,13,14* Abstract Background: Osteoarthritis is a chronic musculoskeletal disorder characterized mainly by progressive degradation of the hyaline cartilage. Patients with osteoarthritis often postpone seeking medical help, which results in the diagnosis being made at an advanced stage of cartilage destruction. Sustained efforts are needed to identify specific markers that might help in early diagnosis, monitoring disease progression and in improving therapeutic outcomes. We employed a multipronged proteomic approach, which included multiple fractionation strategies followed by high resolution mass spectrometry analysis to explore the proteome of synovial fluid obtained from osteoarthritis patients. In addition to the total proteome, we also enriched glycoproteins from synovial fluid using lectin affinity chromatography. Results: We identified 677 proteins from synovial fluid of patients -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Inflammatory Modulation of Hematopoietic Stem Cells by Magnetic Resonance Imaging
Electronic Supplementary Material (ESI) for RSC Advances. This journal is © The Royal Society of Chemistry 2014 Inflammatory modulation of hematopoietic stem cells by Magnetic Resonance Imaging (MRI)-detectable nanoparticles Sezin Aday1,2*, Jose Paiva1,2*, Susana Sousa2, Renata S.M. Gomes3, Susana Pedreiro4, Po-Wah So5, Carolyn Ann Carr6, Lowri Cochlin7, Ana Catarina Gomes2, Artur Paiva4, Lino Ferreira1,2 1CNC-Center for Neurosciences and Cell Biology, University of Coimbra, Coimbra, Portugal, 2Biocant, Biotechnology Innovation Center, Cantanhede, Portugal, 3King’s BHF Centre of Excellence, Cardiovascular Proteomics, King’s College London, London, UK, 4Centro de Histocompatibilidade do Centro, Coimbra, Portugal, 5Department of Neuroimaging, Institute of Psychiatry, King's College London, London, UK, 6Cardiac Metabolism Research Group, Department of Physiology, Anatomy & Genetics, University of Oxford, UK, 7PulseTeq Limited, Chobham, Surrey, UK. *These authors contributed equally to this work. #Correspondence to Lino Ferreira ([email protected]). Experimental Section Preparation and characterization of NP210-PFCE. PLGA (Resomers 502 H; 50:50 lactic acid: glycolic acid) (Boehringer Ingelheim) was covalently conjugated to fluoresceinamine (Sigma- Aldrich) according to a protocol reported elsewhere1. NPs were prepared by dissolving PLGA (100 mg) in a solution of propylene carbonate (5 mL, Sigma). PLGA solution was mixed with perfluoro- 15-crown-5-ether (PFCE) (178 mg) (Fluorochem, UK) dissolved in trifluoroethanol (1 mL, Sigma). This solution was then added to a PVA solution (10 mL, 1% w/v in water) dropwise and stirred for 3 h. The NPs were then transferred to a dialysis membrane and dialysed (MWCO of 50 kDa, Spectrum Labs) against distilled water before freeze-drying. Then, NPs were coated with protamine sulfate (PS).