Crustacea: Decapoda: Brachyura: Potamidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Filariasis and Its Control in Fujian, China
REVIEW FILARIASIS AND ITS CONTROL IN FUJIAN, CHINA Liu ling-yuan, Liu Xin-ji, Chen Zi, Tu Zhao-ping, Zheng Guo-bin, Chen Vue-nan, Zhang Ying-zhen, Weng Shao-peng, Huang Xiao-hong and Yang Fa-zhu Fujian Provincial Institute of Parasitic Diseases, Fuzhou, Fujian, China. Abstract. Epidemiological survey of filariasis in Fujian Province, China showed that malayan filariasis, transmitted by Anopheles lesteri anthropophagus was mainly distributed in the northwest part and bancrof tian filariasis with Culex quinquefasciatus as vector, in middle and south coastal regions. Both species of filariae showed typical nocturnal periodicity. Involvement of the extremities was not uncommon in mala yan filariasis. In contrast, hydrocele was often present in bancroftian filariasis, in which limb impairment did not appear so frequently as in the former. Hetrazan treatment was administered to the microfilaremia cases identified during blood examination surveys, which were integrated with indoor residual spraying of insecticides in endemic areas of malayan filariasis when the vector mosquito was discovered and with mass treatment with hetrazan medicated salt in endemic areas of bancroft ian filariasis. At the same time the habitation condition was improved. These factors facilitated the decrease in incidence. As a result malayan and bancroftian filariasis were proclaimed to have reached the criterion of basic elimination in 1985 and 1987 respectively. Surveillance was pursued thereafter and no signs of resurgence appeared. DISCOVER Y OF FILARIASIS time: he found I male and 16 female adult filariae in retroperitoneallymphocysts and a lot of micro Fujian Province is situated between II S050' filariae in pulmonary capillaries and glomeruli at to 120°43' E and 23°33' to 28°19' N, on the south 8.30 am (Sasa, 1976). -

China in 50 Dishes
C H I N A I N 5 0 D I S H E S CHINA IN 50 DISHES Brought to you by CHINA IN 50 DISHES A 5,000 year-old food culture To declare a love of ‘Chinese food’ is a bit like remarking Chinese food Imported spices are generously used in the western areas you enjoy European cuisine. What does the latter mean? It experts have of Xinjiang and Gansu that sit on China’s ancient trade encompasses the pickle and rye diet of Scandinavia, the identified four routes with Europe, while yak fat and iron-rich offal are sauce-driven indulgences of French cuisine, the pastas of main schools of favoured by the nomadic farmers facing harsh climes on Italy, the pork heavy dishes of Bavaria as well as Irish stew Chinese cooking the Tibetan plains. and Spanish paella. Chinese cuisine is every bit as diverse termed the Four For a more handy simplification, Chinese food experts as the list above. “Great” Cuisines have identified four main schools of Chinese cooking of China – China, with its 1.4 billion people, has a topography as termed the Four “Great” Cuisines of China. They are Shandong, varied as the entire European continent and a comparable delineated by geographical location and comprise Sichuan, Jiangsu geographical scale. Its provinces and other administrative and Cantonese Shandong cuisine or lu cai , to represent northern cooking areas (together totalling more than 30) rival the European styles; Sichuan cuisine or chuan cai for the western Union’s membership in numerical terms. regions; Huaiyang cuisine to represent China’s eastern China’s current ‘continental’ scale was slowly pieced coast; and Cantonese cuisine or yue cai to represent the together through more than 5,000 years of feudal culinary traditions of the south. -

Download Article
Advances in Social Science, Education and Humanities Research, volume 341 5th International Conference on Arts, Design and Contemporary Education (ICADCE 2019) Field Investigation on Eight Tones in Matang Village, Renhua County* Qunying Wang Xiaoyan Chen School of Music School of Music Shaoguan University Shaoguan University Shaoguan, China 512005 Shaoguan, China 512005 Abstract—The Eight-tone Band (also called Matang Drum the above Guangdong folk art forms. It is the "eight-tone Band) of Matang village, Renhua county in Shaoguan city, troupe". The so-called eight-tone music originally refers to Guangdong province is an active local eight-tone club. It has the classification name of ancient Musical Instruments. Here been providing the villagers with performance for happy it refers to a folk music activity, a pure instrumental form. occasions and funeral affairs since the 60s except during the The term "eight tones" first appeared in the Zhou Dynasty. Cultural Revolution. Their music can be divided to music for At that time, instruments were divided into eight categories happy occasions and for funeral affairs, the former being according to the different materials, namely "metal, stone, joyous and cheerful and the latter sad and low-pitched. But clay, leather, silk, wood, gourd, bamboo". Later, it was with the development of the society, the Eight-tone Band is also widely used to refer to musical instruments. Now the changing. generally referred "eight-tone troupe" refers to a kind of rural Keywords—the Eight-tone Band; pattern of manifestation; folk music (including blowing, playing, drumming, singing), music characteristics; status quo of inheritance and "eight-tone music" is the music played by the eight-tone troupes. -

Filed By: [email protected], Filed Date: 1/7/20 11:04 PM, Submission Status: Approved Page 47 of 123 Barcode:3927422-02 A-351-853 INV - Investigation
Barcode:3927422-02 A-351-853 INV - Investigation - Company Name Address E-mail Phone Website Estrada Municipal - CDR 455, S / N | km 1 Castilian 55 49 3561-3248 and 55- Adami S/A Madeiras Caçador (SC) | Postal Code 89514-899 B [email protected] 49-9184-1887 http://www.adami.com.br/ Rua Distrito Industrial - Quadra 06 - lote 03 - Setor D, Advantage Florestal Ananindeua - PA, 67035-330, Brazil [email protected] 55(91) 3017-5565 https://advantageflorestal.com.br/contact-us/ São Josafat, 1850 Street - Clover - Prudentópolis AFFONSO DITZEL & CIA LTDA Paraná - Brazil - ZIP Code 84400-000 [email protected] 55 42 3446-1440 https://www.affonsoditzel.com/index.php AG Intertrade [email protected] 55 41 3015-5002 http://www.agintertrade.com.br/en/home-2/ General Câmara Street, 243/601 55-51-2217-7344 and Araupel SA 90010-230 - Porto Alegre, RS - Brazil [email protected] 55-51-3254-8900 http://www.araupel.com.br/ Rua Félix da Cunha, 1009 – 8º andar CEP: 90570-001 [email protected] and 55 43 3535-8300 and 55- Braspine Madeiras Ltda. Porto Alegre – RS [email protected] 42-3271-3000 http://www.braspine.com.br/en/home/ R. Mal. Floriano Peixoto, 1811 - 12° andar, Sala 124 - Brazil South Lumber Centro, Guarapuava - PR, 85010-250, Brazil [email protected] 55 42 3622-9185 http://brazilsouthlumber.com.br/?lang=en Curupaitis Street, 701 - Curitiba - Paraná - Brazil - ZIP COMERCIAL EXPORTADORA WK LTDA Code 80.310-180 [email protected] http://wktrading.com.br/ 24 de Outubro Street, -

The Spatial Differentiation of the Suitability of Ice-Snow Tourist Destinations Based on a Comprehensive Evaluation Model in China
sustainability Article The Spatial Differentiation of the Suitability of Ice-Snow Tourist Destinations Based on a Comprehensive Evaluation Model in China Jun Yang 1,*, Ruimeng Yang 1, Jing Sun 1, Tai Huang 2,3,* and Quansheng Ge 3 1 Liaoning Key Laboratory of Physical Geography and Geomatics, Liaoning Normal University, Dalian 116029, China; [email protected] (R.Y.); [email protected] (J.S.) 2 Department of Tourism Management, Soochow University, Suzhou 215123, China 3 Key Laboratory of Land Surface Patterns and Simulation, Institute of Geographic Sciences and Natural Resources Research, CAS, Beijing 100101, China; [email protected] * Correspondence: [email protected] (J.Y.); [email protected] (T.H.) Academic Editors: Jun Liu, Gang Liu and This Rutishauser Received: 1 February 2017; Accepted: 4 May 2017; Published: 8 May 2017 Abstract: Ice, snow, and rime are wonders of the cold season in an alpine climate zone and climate landscape. With its pure, spectacular, and magical features, these regions attract numerous tourists. Ice and snow landscapes can provide not only visually-stimulating experiences for people, but also opportunities for outdoor play and movement. In China, ice and snow tourism is a new type of recreation; however, the establishment of snow and ice in relation to the suitability of the surrounding has not been clearly expressed. Based on multi-source data, such as tourism, weather, and traffic data, this paper employs the Delphi-analytic hierarchy process (AHP) evaluation method and a spatial analysis method to study the spatial differences of snow and ice tourism suitability in China. China’s ice and snow tourism is located in the latitude from 35◦N to 53.33◦N and latitude 41.5◦N to 45◦N and longitude 82◦E to 90◦E, with the main focus on latitude and terrain factors. -

The Paradigm of Hakka Women in History
DOI: 10.4312/as.2021.9.1.31-64 31 The Paradigm of Hakka Women in History Sabrina ARDIZZONI* Abstract Hakka studies rely strongly on history and historiography. However, despite the fact that in rural Hakka communities women play a central role, in the main historical sources women are almost absent. They do not appear in genealogy books, if not for their being mothers or wives, although they do appear in some legends, as founders of villages or heroines who distinguished themselves in defending the villages in the absence of men. They appear in modern Hakka historiography—Hakka historiography is a very recent discipline, beginning at the end of the 19th century—for their moral value, not only for adhering to Confucian traditional values, but also for their endorsement of specifically Hakka cultural values. In this paper we will analyse the cultural paradigm that allows women to become part of Hakka history. We will show how ethical values are reflected in Hakka historiography through the reading of the earliest Hakka historians as they depict- ed Hakka women. Grounded on these sources, we will see how the narration of women in Hakka history has developed until the present day. In doing so, it is necessary to deal with some relevant historical features in the construc- tion of Hakka group awareness, namely migration, education, and women narratives, as a pivotal foundation of Hakka collective social and individual consciousness. Keywords: Hakka studies, Hakka woman, women practices, West Fujian Paradigma žensk Hakka v zgodovini Izvleček Študije skupnosti Hakka se močno opirajo na zgodovino in zgodovinopisje. -

Climatic Disasters and Defense Countermeasures of the Oasis City
Climatic Disasters and Defense Countermeasures of the Oasis City on Tropic of Cancer Duan Peng LingZhao JiaFengWeng (Zhaoqing Meteorological Observatory, Guangdong, China 526060) Abstract:This paper analyzes the climatic characteristics and climatic disasters of the oasis city of zhaoqing on the tropic of Cancer .The result indicates that the frequent meteorological droughts, and the frequent Geological disasters caused by heavy rain,and the high temperature,which cause energy consumption and electricity consumption, and the smog, the severe thunderstorms and short-term strong winds which effect on urban transport. And the impact of dominant winds on industrial layout, and some defense countermeasures have been put forward:Ecological city planning should consider meteorological risk areas according to meteorological conditions; Climate demonstration must be conducted for major urban projects;Strengthen the relevant research of meteorological planning for eco-city construction and other countermeasures. These efforts will provide scientific data for the government departments to plan for the sustainable development of ecological cities. Key words: Oasis City; Climate characteristics; Climate disasters;Countermeasure 1.Introduction Zhaoqing City, Guangdong Province is located in the central and western part of Guangdong Province. It is located in the south of Nanling, with high mountains in the Northwest and low in the Southeast. The mountains, hilly basins, river valleys, and plains criss-cross each other. The topography is complex and diverse. The entire territory of Zhaoqing is between 22 ° 47 ′ and 24 ° 24 ′ north latitude, and the Tropic of Cancer runs through it. Due to the subtropical monsoon and monsoon humid climate and the high and low terrain in the Northwest and Southeast, the climate is hot and rainy. -

Volume 90 Number 4 2003 Annals of the Missouri Botanical Garden
Volume 90 Annals Number 4 of the 2003 Missouri Botanical Garden A REVISION OF THE Yelin Huang,2 Peter W. Fritsch,3 and 2 IMBRICATE GROUP OF Suhua Shi STYRAX SERIES CYRTA (STYRACACEAE) IN ASIA1 ABSTRACT Several taxonomic treatments of Styrax (Styracaceae) exist in regional ¯oras of Asia, but the Asian species of the genus have not been comprehensively revised since 1907. To help rectify this, we conducted a taxonomic revision of the Asian species of Styrax series Cyrta with imbricate corolla aestivation. Our revision comprises 17 species with a combined distribution from Japan south to Sumatra and west to Nepal. The circumscriptions of the heretofore poorly de®ned species S. hookeri and S. serrulatus are clari®ed. Styrax agrestis var. curvirostratus is elevated to the species level, and lectotypes are selected for S. duclouxii, S. ¯oribundus, S. hemsleyanus, S. hookeri, S. hookeri var. yunnanensis, S. hypoglaucus, S. japonicus, S. limprichtii, S. macranthus, S. obassia, S. perkinsiae, S. serrulatus var. latifolius, S. shiraianus, S. supaii, and S. wilsonii. Keys, descriptions, and distribution maps are provided for all species. Key words: eastern Asia, Styracaceae, Styrax, Styrax series Cyrta. Styrax L. comprises about 130 species of trees ern Argentina and Uruguay (Fritsch, 1999, 2001). and shrubs distributed in eastern and southeastern Styrax is by far the largest and most widespread of Asia, the New World, and the Mediterranean region the 11 genera in the Styracaceae sensu Fritsch et (Fritsch, 1999). The range of this genus is typical al. (2001) and Fritsch (in press a). Characters of many plant groups distributed among the refugia unique to Styrax in relation to the family include a of Tertiary mixed-mesophytic forests in the North- stamen tube attached high (vs. -
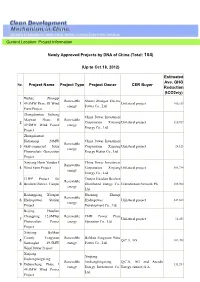
Up to Oct 18, 2012)
Current Location: Project Information Newly Approved Projects by DNA of China (Total: 104) (Up to Oct 18, 2012) Estimated Ave. GHG No. Project Name Project Type Project Owner CER Buyer Reduction (tCO2e/y) Wulate Zhongqi Renewable Shanxi Zhangze Electric 1 49.5MW Phase III Wind Unilateral project 106,339 energy Power Co., Ltd. Farm Project Zhongdiantou Tacheng China Power Investment Mayitasi Phase II Renewable 2 Corporation Xinjiang Unilateral project 110,917 49.5MW Wind Power energy Energy Co., Ltd. Project Zhongdiantou Hetianerqi 20MW China Power Investment Renewable 3 Grid-connected Solar Corporation Xinjiang Unilateral project 24,121 energy Photovoltaic Generation Energy Hetian Co., Ltd. Project Xinjiang Hami Yandun I China Power Investment Renewable 4 Wind Farm Project Corporation Xinjiang Unilateral project 386,734 energy Energy Co., Ltd. CHPP Project for Tianjin Huadian Beichen Renewable 5 Beichen District, Tianjin Distributed Energy Co., GreenStream Network Plc 208,064 energy Ltd. Bailongjiang Xi'ergou Huaneng Zhouqu Renewable 6 Hydropower Station Hydropower Unilateral project 247,663 energy Project Development Co., Ltd. Beijing Huadian Changping 12.8MWp Renewable CHD Power Plant 7 Unilateral project 14,455 Photovoltaic Power energy Operation Co., Ltd. Project Xinjiang Balikun County Fengyuan Renewable Balikun Fengyuan Wind 8 Q.C.A. AG 102,498 Santanghu 49.5MW energy Power Co., Ltd. Wind Power Project Xinjiang Xinjiang Jinshangfengxiang Renewable Jinshangfengxiang Q.C.A. AG and Arcadia 9 Dabancheng Phase I 111,284 energy Energy Investment Co., Energy (Suisse) S.A. 49.5MW Wind Power Ltd. Project Gansu Jinchuan 50MW Jinchang Jintai Solar Photovoltaic Renewable 10 Photovoltaic Power Co., Unilateral project 55,115 Power Generation energy Ltd. -

Canton Confederation
DISCLAIMER: The Topps Company, Inc. has sole ownership of the names, and/or any proprietary material used in connection with the game Shadowrun. The Topps Company, Inc. is not affiliated with the author, Robert Derie, in any official capacity whatsoever. The information contained in this document is for non-commercial entertainment purposes only. CATO COFEDERATIO >>>> BEGI FACTS AD DEMOGRAPHIC BOX Canton Confederation: Fujian (Fu-chen) , Fuzhou (Fu-chow), Guangdong (Can-ton) , Guangzhou (Can-chow), Jiangxi (Chiang-zi) Prominent Languages: Yue (Cantonese), Mandarin, Xiang, Pinghua Population: 220,560,000 List of Provinces: Fujian, Guangdong, Hunan, Jiangxi, Macao Government Type: Confederation of democratic republics Bordering Countries: Coastal Provinces, Guangxi, Henan, Honk Kong, Sichuan, Taiwan Geography: The southern part of the Canton Confederation is dominated by the Pearl River and its extensive coastline with the South China Sea; it is separated from the northern territories by the Nanling Mountains. The northern provinces are much more hilly and mountainous, featuring several small mountain ranges before flattening out again around Lake Poyang, the largest freshwater lake in the Chinese Splinter States. otable Features: Pearl River Delta, South China Sea Territories, Lake Poyang, Zhurong Peak >>>> ED FACTS AD DEMOGRAPHICS BOX GEOPOLITICAL OVERVIEW In 2018, the members of the Greater Canton Economic Development Council, a trade alliance of the most industrially developed provinces (Fujian, Guangdong, Hunan, Jiangxi, and Zhejiang), seceded from the People’s Republic of China and formed an independent confederation, sparking further secession movements and leading to the Republic Civil War (2019-2027), which ended with a fractured collection of Chinese states. When secession was declared, the Canton Confederation invited Taiwan to join them, but the state declined. -

Table of Codes for Each Court of Each Level
Table of Codes for Each Court of Each Level Corresponding Type Chinese Court Region Court Name Administrative Name Code Code Area Supreme People’s Court 最高人民法院 最高法 Higher People's Court of 北京市高级人民 Beijing 京 110000 1 Beijing Municipality 法院 Municipality No. 1 Intermediate People's 北京市第一中级 京 01 2 Court of Beijing Municipality 人民法院 Shijingshan Shijingshan District People’s 北京市石景山区 京 0107 110107 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Haidian District of Haidian District People’s 北京市海淀区人 京 0108 110108 Beijing 1 Court of Beijing Municipality 民法院 Municipality Mentougou Mentougou District People’s 北京市门头沟区 京 0109 110109 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Changping Changping District People’s 北京市昌平区人 京 0114 110114 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Yanqing County People’s 延庆县人民法院 京 0229 110229 Yanqing County 1 Court No. 2 Intermediate People's 北京市第二中级 京 02 2 Court of Beijing Municipality 人民法院 Dongcheng Dongcheng District People’s 北京市东城区人 京 0101 110101 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Xicheng District Xicheng District People’s 北京市西城区人 京 0102 110102 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Fengtai District of Fengtai District People’s 北京市丰台区人 京 0106 110106 Beijing 1 Court of Beijing Municipality 民法院 Municipality 1 Fangshan District Fangshan District People’s 北京市房山区人 京 0111 110111 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Daxing District of Daxing District People’s 北京市大兴区人 京 0115 -

Peer Reviewed Title: Critical Han Studies: the History, Representation, and Identity of China's Majority Author: Mullaney, Thoma
Peer Reviewed Title: Critical Han Studies: The History, Representation, and Identity of China's Majority Author: Mullaney, Thomas S. Leibold, James Gros, Stéphane Vanden Bussche, Eric Editor: Mullaney, Thomas S.; Leibold, James; Gros, Stéphane; Vanden Bussche, Eric Publication Date: 02-15-2012 Series: GAIA Books Permalink: http://escholarship.org/uc/item/07s1h1rf Keywords: Han, Critical race studies, Ethnicity, Identity Abstract: Addressing the problem of the ‘Han’ ethnos from a variety of relevant perspectives—historical, geographical, racial, political, literary, anthropological, and linguistic—Critical Han Studies offers a responsible, informative deconstruction of this monumental yet murky category. It is certain to have an enormous impact on the entire field of China studies.” Victor H. Mair, University of Pennsylvania “This deeply historical, multidisciplinary volume consistently and fruitfully employs insights from critical race and whiteness studies in a new arena. In doing so it illuminates brightly how and when ideas about race and ethnicity change in the service of shifting configurations of power.” David Roediger, author of How Race Survived U.S. History “A great book. By examining the social construction of hierarchy in China,Critical Han Studiessheds light on broad issues of cultural dominance and in-group favoritism.” Richard Delgado, author of Critical Race Theory: An Introduction “A powerful, probing account of the idea of the ‘Han Chinese’—that deceptive category which, like ‘American,’ is so often presented as a natural default, even though it really is of recent vintage. A feast for both Sinologists and comparativists everywhere.” Magnus Fiskesjö, Cornell University eScholarship provides open access, scholarly publishing services to the University of California and delivers a dynamic research platform to scholars worldwide.