Implications for the Facio-Scapulo-Humeral Dystrophy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FSHD Region Gene 1 (FRG1) Is Crucial for Angiogenesis Linking FRG1 to Facioscapulohumeral Muscular Dystrophy-Associated Vasculopathy
University of Massachusetts Medical School eScholarship@UMMS Peter Jones Lab Publications Cell and Developmental Biology Laboratories 2009-05-01 FSHD region gene 1 (FRG1) is crucial for angiogenesis linking FRG1 to facioscapulohumeral muscular dystrophy-associated vasculopathy Ryan Wuebbles University of Illinois at Urbana-Champaign Et al. Let us know how access to this document benefits ou.y Follow this and additional works at: https://escholarship.umassmed.edu/peterjones Part of the Cell Biology Commons, Developmental Biology Commons, Molecular Biology Commons, Molecular Genetics Commons, Musculoskeletal Diseases Commons, and the Nervous System Diseases Commons Repository Citation Wuebbles R, Hanel ML, Jones PL. (2009). FSHD region gene 1 (FRG1) is crucial for angiogenesis linking FRG1 to facioscapulohumeral muscular dystrophy-associated vasculopathy. Peter Jones Lab Publications. https://doi.org/10.1242/dmm.002261. Retrieved from https://escholarship.umassmed.edu/ peterjones/11 Creative Commons License This work is licensed under a Creative Commons Attribution 3.0 License. This material is brought to you by eScholarship@UMMS. It has been accepted for inclusion in Peter Jones Lab Publications by an authorized administrator of eScholarship@UMMS. For more information, please contact [email protected]. Disease Models & Mechanisms 2, 267-274 (2009) doi:10.1242/dmm.002261 RESEARCH ARTICLE FSHD region gene 1 (FRG1) is crucial for angiogenesis linking FRG1 to facioscapulohumeral muscular dystrophy-associated vasculopathy Ryan D. Wuebbles1,*, Meredith L. Hanel1,* and Peter L. Jones1,‡ SUMMARY The genetic lesion that is diagnostic for facioscapulohumeral muscular dystrophy (FSHD) results in an epigenetic misregulation of gene expression, which ultimately leads to the disease pathology. FRG1 (FSHD region gene 1) is a leading candidate for a gene whose misexpression might lead to FSHD. -

Gene Expression During Normal and FSHD Myogenesis Tsumagari Et Al
Gene expression during normal and FSHD myogenesis Tsumagari et al. Tsumagari et al. BMC Medical Genomics 2011, 4:67 http://www.biomedcentral.com/1755-8794/4/67 (27 September 2011) Tsumagari et al. BMC Medical Genomics 2011, 4:67 http://www.biomedcentral.com/1755-8794/4/67 RESEARCHARTICLE Open Access Gene expression during normal and FSHD myogenesis Koji Tsumagari1, Shao-Chi Chang1, Michelle Lacey2,3, Carl Baribault2,3, Sridar V Chittur4, Janet Sowden5, Rabi Tawil5, Gregory E Crawford6 and Melanie Ehrlich1,3* Abstract Background: Facioscapulohumeral muscular dystrophy (FSHD) is a dominant disease linked to contraction of an array of tandem 3.3-kb repeats (D4Z4) at 4q35. Within each repeat unit is a gene, DUX4, that can encode a protein containing two homeodomains. A DUX4 transcript derived from the last repeat unit in a contracted array is associated with pathogenesis but it is unclear how. Methods: Using exon-based microarrays, the expression profiles of myogenic precursor cells were determined. Both undifferentiated myoblasts and myoblasts differentiated to myotubes derived from FSHD patients and controls were studied after immunocytochemical verification of the quality of the cultures. To further our understanding of FSHD and normal myogenesis, the expression profiles obtained were compared to those of 19 non-muscle cell types analyzed by identical methods. Results: Many of the ~17,000 examined genes were differentially expressed (> 2-fold, p < 0.01) in control myoblasts or myotubes vs. non-muscle cells (2185 and 3006, respectively) or in FSHD vs. control myoblasts or myotubes (295 and 797, respectively). Surprisingly, despite the morphologically normal differentiation of FSHD myoblasts to myotubes, most of the disease-related dysregulation was seen as dampening of normal myogenesis- specific expression changes, including in genes for muscle structure, mitochondrial function, stress responses, and signal transduction. -

SNF2 Chromatin Remodeler-Family Proteins FRG1 and -2 Are Required for RNA-Directed DNA Methylation
SNF2 chromatin remodeler-family proteins FRG1 and -2 are required for RNA-directed DNA methylation Martin Grotha, Hume Strouda,1, Suhua Fenga,b,c, Maxim V. C. Greenberga,2, Ajay A. Vashishtd, James A. Wohlschlegeld, Steven E. Jacobsena,b,c,3, and Israel Ausine,3 aDepartment of Molecular, Cell, and Developmental Biology, bEli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research, dDepartment of Biological Chemistry, David Geffen School of Medicine, cHoward Hughes Medical Institute, University of California, Los Angeles, CA 90095; and eBasic Forestry and Biotechnology Center, Fujian Agriculture and Forestry University, Fujian, Fuzhou 350002, China Contributed by Steven E. Jacobsen, October 29, 2014 (sent for review August 2, 2014) DNA methylation in Arabidopsis thaliana is maintained by at least four (6, 7). Recruitment of Pol V is mediated by methyl-CG–binding different enzymes: DNA METHYLTRANSFERASE1 (MET1), CHROMO- noncatalytic Su(var)3-9 histone methyltransferase homologs METHYLASE3 (CMT3), DOMAINS REARRANGED METHYLTRANSFER- SUVH2 and SUVH9, and a putative chromatin remodeling ASE2 (DRM2), and CHROMOMETHYLASE2 (CMT2). However, DNA complex called the DRD1-DMS3-RDM1 complex (8, 9). In ad- methylation is established exclusively by the enzyme DRM2, which dition to SUVH2 and SUVH9, SUVR2 from the same family of acts in the RNA-directed DNA methylation (RdDM) pathway. Some SET-domain proteins was also identified as an RdDM factor by RdDM components belong to gene families and have partially redun- a systematic analysis of DNA methylation defects in mutants of dant functions, such as the endoribonucleases DICER-LIKE 2, 3,and4, Su(var)3-9 homologs (10). However, the precise molecular function and INVOLVED IN DE NOVO2 (IDN2) interactors IDN2-LIKE 1 and 2. -

The DNA Sequence and Comparative Analysis of Human Chromosome 20
articles The DNA sequence and comparative analysis of human chromosome 20 P. Deloukas, L. H. Matthews, J. Ashurst, J. Burton, J. G. R. Gilbert, M. Jones, G. Stavrides, J. P. Almeida, A. K. Babbage, C. L. Bagguley, J. Bailey, K. F. Barlow, K. N. Bates, L. M. Beard, D. M. Beare, O. P. Beasley, C. P. Bird, S. E. Blakey, A. M. Bridgeman, A. J. Brown, D. Buck, W. Burrill, A. P. Butler, C. Carder, N. P. Carter, J. C. Chapman, M. Clamp, G. Clark, L. N. Clark, S. Y. Clark, C. M. Clee, S. Clegg, V. E. Cobley, R. E. Collier, R. Connor, N. R. Corby, A. Coulson, G. J. Coville, R. Deadman, P. Dhami, M. Dunn, A. G. Ellington, J. A. Frankland, A. Fraser, L. French, P. Garner, D. V. Grafham, C. Grif®ths, M. N. D. Grif®ths, R. Gwilliam, R. E. Hall, S. Hammond, J. L. Harley, P. D. Heath, S. Ho, J. L. Holden, P. J. Howden, E. Huckle, A. R. Hunt, S. E. Hunt, K. Jekosch, C. M. Johnson, D. Johnson, M. P. Kay, A. M. Kimberley, A. King, A. Knights, G. K. Laird, S. Lawlor, M. H. Lehvaslaiho, M. Leversha, C. Lloyd, D. M. Lloyd, J. D. Lovell, V. L. Marsh, S. L. Martin, L. J. McConnachie, K. McLay, A. A. McMurray, S. Milne, D. Mistry, M. J. F. Moore, J. C. Mullikin, T. Nickerson, K. Oliver, A. Parker, R. Patel, T. A. V. Pearce, A. I. Peck, B. J. C. T. Phillimore, S. R. Prathalingam, R. W. Plumb, H. Ramsay, C. M. -

Human Proteins That Interact with RNA/DNA Hybrids
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Resource Human proteins that interact with RNA/DNA hybrids Isabel X. Wang,1,2 Christopher Grunseich,3 Jennifer Fox,1,2 Joshua Burdick,1,2 Zhengwei Zhu,2,4 Niema Ravazian,1 Markus Hafner,5 and Vivian G. Cheung1,2,4 1Howard Hughes Medical Institute, Chevy Chase, Maryland 20815, USA; 2Life Sciences Institute, University of Michigan, Ann Arbor, Michigan 48109, USA; 3Neurogenetics Branch, National Institute of Neurological Disorders and Stroke, NIH, Bethesda, Maryland 20892, USA; 4Department of Pediatrics, University of Michigan, Ann Arbor, Michigan 48109, USA; 5Laboratory of Muscle Stem Cells and Gene Regulation, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Bethesda, Maryland 20892, USA RNA/DNA hybrids form when RNA hybridizes with its template DNA generating a three-stranded structure known as the R-loop. Knowledge of how they form and resolve, as well as their functional roles, is limited. Here, by pull-down assays followed by mass spectrometry, we identified 803 proteins that bind to RNA/DNA hybrids. Because these proteins were identified using in vitro assays, we confirmed that they bind to R-loops in vivo. They include proteins that are involved in a variety of functions, including most steps of RNA processing. The proteins are enriched for K homology (KH) and helicase domains. Among them, more than 300 proteins preferred binding to hybrids than double-stranded DNA. These proteins serve as starting points for mechanistic studies to elucidate what RNA/DNA hybrids regulate and how they are regulated. -

Characterization of C. Elegans Pat-9 and Frg-1, Genes Critical for Body Wall Muscle Development
CHARACTERIZATION OF C. ELEGANS PAT-9 AND FRG-1, GENES CRITICAL FOR BODY WALL MUSCLE DEVELOPMENT BY QIAN LIU DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Cell and Developmental Biology in the Graduate College of the University of Illinois at Urbana-Champaign, 2010 Urbana, Illinois Doctoral Committee Assistant Professor Peter L. Jones, Director of Research Associate Professor Jie Chen, Chair Professor Philip M. Best Associate Professor Phillip A. Newmark Assistant Professor William M. Brieher Abstract Caenorhabditis elegans, a small free-living nematode, has been used to investigate many biological processes including apoptosis, the cell cycle, gene regulation, cell polarity, metabolism, aging, and development. Transgenic C. elegans have been broadly utilized as a human disease model to study mechanisms of pathology for diseases such as obesity, diabetes, cancer, Duchenne’s muscular dystrophy, and several neurodegenerative diseases. In these studies we used C. elegans to further investigate muscle development and as a model for facioscapulohumeral muscular dystrophy (FSHD) pathophysiology. A previous genetic screen in C. elegans investigating early muscle development isolated mutants producing a pat (paralyzed, elongation arrested at two-fold) phenotype. This project focused on identifying and characterizing one of the pat genes, pat-9. Positional cloning and reverse genetics were used to identify T27b1.2 as encoding the pat-9 gene. Characterization of the PAT-9 protein revealed that it is an exclusively nuclear zinc finger protein required for muscle development. This nuclear localization for PAT-9 makes it unique among the pat family of genes. Considering the pat-9 mutant phenotype shows severely disrupted muscle attachment sites despite PAT-9 not localizing to those sites, PAT-9 may function in the regulation of expression for some necessary structural component of the muscle attachment sites. -

A Possible Approach for Treating FSHD with Rnai Therapeutics 4 10 16 24
A Publication of the Facioscapulohumeral Muscular Dystrophy Society SUMMER 2011 RESEARCH ISSUE FSH Watch 2011 CONNECTING THE COMMUNITY OF PATIENTS, FAMILIES, CLINICIANS AND INVESTIGATORS TRANSLATIONAL RESEArcH Journey toward developing a drug for FSHD Perspectives and updates from recently funded FSH Society grantees by DARKO BOSNAKOVSKI, D.V.M., Ph.D. University Goce Delcev Stip, Macedonia lthough FSHD is considered one Aof the most common inherited neuromuscular diseases, there is no Dr. Davide Gabellini with fellow researchers at the 2010 FSH Society FSHD research workshop specific therapeutic practice for it. So what can we do about it? First, we have TRANSLATIONAL RESEArcH to understand the mechanisms of the disease and to identify the crucial links in the chain of molecular reactions A possible approach for treating FSHD that underline FSHD. Furthermore, we have to develop a system to screen with RNAi therapeutics various therapeutic approaches, and in Perspectives and updates from FSH Society grantees the end to generate an animal model where clinical relevance of therapy by DANIEL PEREZ scientists: the Harper Lab at The Ohio State can be determined. When I joined the FSH Society University and Nationwide Children’s Hos- FSHD group lead by Dr. Michael Kyba with contributions by DAVIDE GABELLINI, Ph.D. pital in Columbus, Ohio, with a collabo- at University of Texas Southwestern six Division of Regenerative Medicine, San Raffaele rator in Modena, Italy; and the Gabellini years ago to develop a specific therapy Scientific Institute, Milan, Italy, and and Chamberlain labs in Milan, Italy, and for FSHD all of the above was considered Seattle, Washington, respectively. -

1 Gene Therapy for Facioscapulohumeral Muscular
Gene Therapy for Facioscapulohumeral Muscular Dystrophy Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Lindsay M. Wallace, B.A. Molecular, Cellular, and Developmental Biology Graduate Program The Ohio State University 2012 Dissertation Committee: Dr. Scott Q. Harper, Advisor Dr. K. Reed Clark Dr. Denis C. Guttridge Dr. Jill Rafael-Fortney 1 Copyright by Lindsay M. Wallace 2012 ABSTRACT Facioscapulohumeral muscular dystrophy is the most common inherited muscular dystrophy though no treatment exists. The lack of therapeutic development for FSHD is directly linked to insufficient understanding of how the disease is caused. The goals of the studies presented here were to gain a better understanding of the pathogenic mechanisms of FSHD and to develop targeted translational therapies to treat the disease. FSHD is associated with D4Z4 repeat contraction on human chromosome 4q35, which does not result in complete loss or mutation of any gene. Consequently, the major obstacle to discerning the underlying pathogenic mechanism is to identify the cause. Although no gene was conclusively linked to FSHD development, evidence supported a role for the D4Z4-encoded DUX4 gene. In Chapter 3, our objective was to test the in vivo myopathic potential of DUX4. We delivered DUX4 to zebrafish and mouse muscle by transposon-mediated transgenesis and adeno-associated viral vectors, respectively. We found over-expression of DUX4 caused abnormalities associated with muscular dystrophy in both animal models. This toxicity required DNA binding, since a DUX4 DNA binding domain mutant produced no abnormalities. We also found the toxic effects of DUX4 were p53-dependent. -
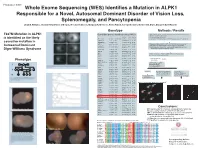
T237M Mutation in ALPK1 Is Identified As the Likely Causative Mutation In
Program #: 3367 Whole Exome Sequencing (WES) Identifies a Mutation in ALPK1 Responsible for a Novel, Autosomal Dominant Disorder of Vision Loss, Splenomegaly, and Pancytopenia Lloyd B. Williams, Chad D. Huff, Denise J. Morgan, Rosann Robinson, Margaux A. Morrison, Krista Kinard, George Rodgers, Kathleen B. Digre, Margaret M. DeAngelis Genotype Methods / Results WES on 4 individuals - 2 affected II-3 III-2, and 2 unaffected II-2 and III-3 T237M Mutation in ALPK1 Candi date genes identified using VAAST Illuimina Truseq Enrichment Kit position Allele Protein Quantitative PCR enrichment is identified as the likely Gene name p-value chromosome (hg19) change change Sequencing with Illumina HiSeq 2000 101 cycle paired end sequencing PRAMEF11 6.10E-06 chr1 12887174 C->T R->H causative mutation in ANKRD20A4 7.32E-06 chr9 69423637 G->A E->K MRPL4 8.55E-06 chr19 10367459 C->T R->W Mapped and aligned with Picard Tools (http://picard.sourceforge.net) FAM90A10 9.77E-06 chr8 7629232 G->T A->S SNPs and indels identified with Genome analysis toolkit (GATK) Autosomal Dominant Manual inspection and curation was done with Intergrative Genomics GOLGA6L10 9.77E-06 chr15 82635194 T->C E->G Viewer (http://www.broadinstitute.org/igv) Digre-Williams Syndrome FAM90A20 1.28E-05 chr8 7155458 C->G A->G EEF1A1 1.65E-05 chr6 74228474 C->T R->H MSI1 1.65E-05 chr12 120800875 C->T V->M VAAST - compares to HGMD, dbSNP, ESP, and1000 Genomes VDAC2 1.71E-05 chr10 76980685 G->T A->S panels to rule out common variants. PIM1 2.08E-05 chr6 37138779 A->T K->M USP11 2.26E-05 chrX -

SORBS2 Transcription Is Activated by Telomere Position Effect-Over Long
Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press SORBS2 Transcription is Activated by Telomere Position Effect-Over Long Distance Upon Telomere Shortening in Muscle Cells from Patients with Facioscapulohumeral Dystrophy Jérôme D. Robin1,4 Andrew T. Ludlow1 Kimberly Batten1 Marie-Cecile Gaillard2; Guido Stadler1; Frédérique Magdinier2; Woodring E. Wright1; Jerry W. Shay1,3,4 1 Department of Cell Biology, UT Southwestern Medical Center, Dallas TX 75390 U.S.A. 2 Aix Marseille Universite, INSERM, GMGF, UMRS 910. 27 Bd Jean Moulin, Marseille 13385 Cedex 05 France. 3 Center for Excellence in Genomics Medicine Research, King Abdulaziz University, Jeddah, Saudi Arabia 4 Co-Corresponding Authors: Jerry W. Shay Tel: 214-648-4201 Fax: 214-648-5814 [email protected] Jérôme D. Robin IRCAN CNRS UMR 7284 / INSERM U1081 Faculté de Médecine 28 avenue de Valombrose 06107 Nice Cedex 2 [email protected] Keywords: chromatin, 4q35 locus, D4Z4, skeletal muscle, myoblast, Hi-C, Telomere, Facioscapulohumeral Dystrophy Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract DNA is organized into complex three-dimensional chromatin structures but how this special organization regulates gene expression remains a central question. These DNA/chromatin looping structures can range in size from 10-20 kilobases (enhancers/repressors) to many megabases (Mb) during intra- and inter- chromosomal interactions. Recently, the influence of telomere length on chromatin organization prior to senescence has revealed the existence of long distance chromatin loops that dictate the expression of genes located up to 10Mb from the telomeres (Telomere Position Effect-Over Long Distances; TPE-OLD). -

Molecular Characterization of Astrocytoma Progression Towards Secondary Glioblastomas Utilizing Patient-Matched Tumor Pairs
cancers Article Molecular Characterization of Astrocytoma Progression Towards Secondary Glioblastomas Utilizing Patient-Matched Tumor Pairs Michael Seifert 1,2,* , Gabriele Schackert 2,3,4, Achim Temme 2,3,4, Evelin Schröck 2,4,5,6,7, Andreas Deutsch 8 and Barbara Klink 2,4,5,9 1 Institute for Medical Informatics and Biometry (IMB), Carl Gustav Carus Faculty of Medicine, Technische Universität Dresden, D-01307 Dresden, Germany 2 National Center for Tumor Diseases (NCT), Partner Site Dresden, D-01307 Dresden, Germany; [email protected] (G.S.); [email protected] (A.T.); [email protected] (E.S.); [email protected] (B.K.) 3 Department of Neurosurgery, Section Experimental Neurosurgery/Tumor Immunology, University Hospital Carl Gustav Carus, Technische Universität Dresden, D-01307 Dresden, Germany 4 German Cancer Consortium (DKTK), Dresden, German Cancer Research Center (DKFZ), D-69120 Heidelberg, Germany 5 Institute for Clinical Genetics, Carl Gustav Carus Faculty of Medicine, Technische Universität Dresden, D-01307 Dresden, Germany 6 ERN-GENTURIS, Hereditary Cancer Syndrome Center Dresden, D-01307 Dresden, Germany 7 Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG), D-01307 Dresden, Germany 8 Center for Information Services and High Performance Computing (ZIH), Technische Universität Dresden, D-01062 Dresden, Germany; [email protected] 9 National Center of Genetics (NCG), Laboratoire national de santé (LNS), L-3555 Dudelange, Luxembourg * Correspondence: [email protected] Received: 19 May 2020; Accepted: 21 June 2020; Published: 26 June 2020 Abstract: Astrocytomas are primary human brain tumors including diffuse or anaplastic astrocytomas that develop towards secondary glioblastomas over time. -

Facioscapulohumeral Muscular Dystrophy and DUX4: Breaking the Silence
Review Facioscapulohumeral muscular dystrophy and DUX4: breaking the silence Silve` re M. van der Maarel1, Rabi Tawil2 and Stephen J. Tapscott3 1 Department of Human Genetics, Leiden University Medical Center, Albinusdreef 2, 2333 ZA, Leiden, Netherlands 2 Department of Neurology, University of Rochester Medical Center, PO Box 673, 601 Elmwood Avenue, Rochester, NY 14642 USA 3 Division of Human Biology, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109 USA Autosomal dominant facioscapulohumeral muscular Figure 1) [6]. The D4Z4 macrosatellite repeat is located dystrophy (FSHD) has an unusual pathogenic mecha- approximately 40–60 kb proximal to the telomere repeat nism. FSHD is caused by deletion of a subset of D4Z4 and varies between 11 and >100 copies of D4Z4 units in the macrosatellite repeat units in the subtelomere of chro- unaffected population [6,7]. The D4Z4 repeat unit is de- mosome 4q. Recent studies provide compelling evi- fined as a 3.3 kb KpnI fragment, and multiple units are dence that a retrotransposed gene in the D4Z4 repeat, ordered head-to-tail to form the D4Z4 repeat array. Most DUX4, is expressed in the human germline and then patients with FSHD1 have a partial and internal deletion epigenetically silenced in somatic tissues. In FSHD, the of the repeat array leaving only 1–10 units on one of their combination of inefficient chromatin silencing of the chromosomes 4. It is believed that at least one unit of D4Z4 D4Z4 repeat and polymorphisms on the FSHD-permis- is necessary to develop FSHD, as monosomy of 4q does not sive alleles that stabilize the DUX4 mRNAs emanating cause FSHD [8].