Physiological Role of Prolylcarboxypeptidase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genome-Wide DNA Methylation Map of Human Neutrophils Reveals Widespread Inter-Individual Epigenetic Variation
www.nature.com/scientificreports OPEN Genome-wide DNA methylation map of human neutrophils reveals widespread inter-individual Received: 15 June 2015 Accepted: 29 October 2015 epigenetic variation Published: 27 November 2015 Aniruddha Chatterjee1,2, Peter A. Stockwell3, Euan J. Rodger1, Elizabeth J. Duncan2,4, Matthew F. Parry5, Robert J. Weeks1 & Ian M. Morison1,2 The extent of variation in DNA methylation patterns in healthy individuals is not yet well documented. Identification of inter-individual epigenetic variation is important for understanding phenotypic variation and disease susceptibility. Using neutrophils from a cohort of healthy individuals, we generated base-resolution DNA methylation maps to document inter-individual epigenetic variation. We identified 12851 autosomal inter-individual variably methylated fragments (iVMFs). Gene promoters were the least variable, whereas gene body and upstream regions showed higher variation in DNA methylation. The iVMFs were relatively enriched in repetitive elements compared to non-iVMFs, and were associated with genome regulation and chromatin function elements. Further, variably methylated genes were disproportionately associated with regulation of transcription, responsive function and signal transduction pathways. Transcriptome analysis indicates that iVMF methylation at differentially expressed exons has a positive correlation and local effect on the inclusion of that exon in the mRNA transcript. Methylation of DNA is a mechanism for regulating gene function in all vertebrates. It has a role in gene silencing, tissue differentiation, genomic imprinting, chromosome X inactivation, phenotypic plasticity, and disease susceptibility1,2. Aberrant DNA methylation has been implicated in the pathogenesis of sev- eral human diseases, especially cancer3–5. Variation in DNA methylation patterns in healthy individuals has been hypothesised to alter human phenotypes including susceptibility to common diseases6 and response to drug treatments7. -

Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps Sinensis on the Treatment of Diabetic Nephropathy
Hindawi Journal of Diabetes Research Volume 2021, Article ID 8891093, 12 pages https://doi.org/10.1155/2021/8891093 Research Article Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps sinensis on the Treatment of Diabetic Nephropathy Yan Li,1 Lei Wang,2 Bojun Xu,1 Liangbin Zhao,1 Li Li,1 Keyang Xu,3 Anqi Tang,1 Shasha Zhou,1 Lu Song,1 Xiao Zhang,1 and Huakui Zhan 1 1Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072 Sichuan, China 2Key Laboratory of Chinese Internal Medicine of Ministry of Education and Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing 100700, China 3Zhejiang Chinese Medical University, Hangzhou, 310053 Zhejiang, China Correspondence should be addressed to Huakui Zhan; [email protected] Received 27 August 2020; Revised 17 January 2021; Accepted 24 January 2021; Published 8 February 2021 Academic Editor: Michaelangela Barbieri Copyright © 2021 Yan Li et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Diabetic nephropathy (DN) is one of the most common complications of diabetes mellitus and is a major cause of end- stage kidney disease. Cordyceps sinensis (Cordyceps, Dong Chong Xia Cao) is a widely applied ingredient for treating patients with DN in China, while the molecular mechanisms remain unclear. This study is aimed at revealing the therapeutic mechanisms of Cordyceps in DN by undertaking a network pharmacology analysis. Materials and Methods. In this study, active ingredients and associated target proteins of Cordyceps sinensis were obtained via Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and Swiss Target Prediction platform, then reconfirmed by using PubChem databases. -

PRCP) As a New Target for Obesity Treatment
Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy Dovepress open access to scientific and medical research Open Access Full Text Article REVIEW Prolylcarboxypeptidase (PRCP) as a new target for obesity treatment B Shariat-Madar1 Abstract: Recently, we serendipitously discovered that mice with the deficiency of the enzyme D Kolte2 prolylcarboxypeptidase (PRCP) have elevated α-melanocyte-stimulating hormone (α-MSH) A Verlangieri2 levels which lead to decreased food intake and weight loss. This suggests that PRCP is an Z Shariat-Madar2 endogenous inactivator of α-MSH and an appetite stimulant. Since a modest weight loss can have the most profound influence on reducing cardiovascular risk factors, the inhibitors of PRCP 1College of Literature, Science, and would be emerging as a possible alternative for pharmacotherapy in high-risk patients with the Arts, University of Michigan, Ann Arbor MI, USA; 2School obesity and obesity-related disorders. The discovery of a new biological activity of PRCP in the of Pharmacy, Department PRCP-deficient mice and studies of α-MSH function indicate the importance and complexity of Pharmacology, University of Mississippi, University, of the hypothalamic pro-opiomelanocortin (POMC) system in altering food intake. Identifying MS, USA a role for PRCP in regulating α-MSH in the brain may be a critical step in enhancing our understanding of how the brain controls food intake and body weight. In light of recent findings, the potential role of PRCP in regulating fuel homeostasis is critically evaluated. Further studies of the role of PRCP in obesity are much needed. Keywords: prolylcarboxypeptidase, melanocyte-stimulating harmone, appetite, weight loss, cardiovascular risk, obesity Introduction The hypothalamus is often times a target for newer potential obesity treatments due to its crucial role in food intake and metabolism. -

Supplemental Material Alternative Splicing Modulated by Genetic
Supplemental Material Alternative Splicing Modulated by Genetic Variants Demonstrates Accelerated Evolution Regulated by Highly Conserved Proteins By Hsiao et al. Supplemental Methods Supplemental References Supplemental Figures 1-8 Supplemental Tables 1-10 (in Excel file) Supplemental Methods RNA-Seq data and mapping Paired-end RNA-Seq data (2x76nt) from seven human cell lines (GM12878, K562, HeLa, HepG2, HUVEC, NHEK, and H1-hESC) were downloaded from the ENCODE data repository (www.encodeproject.org) under the ENCODE Data Coordination Center accession number ENCSR037HRJ, or NCBI GEO accession number GSE30567, where two biological replicates are available for all cell lines except H1-hESC. The reads were mapped using a stringent mapping method described in our previous work (Bahn et al. 2012). Briefly, each end of the paired-end reads was mapped to the hg19 genome and transcriptome using BLAT (Kent 2002) and Bowtie (Langmead et al. 2009), allowing 12 mismatches in each read to collect as many mapping positions as possible. The following mapping parameters were used: BLAT (version 3.4): -minIdentity=75 -tileSize=11; Bowtie (version 0.12.3): -k 80 -e 140 -n 3 -l 20. After initial mapping, we checked for proper pairing with the correct orientations and distance of at most 500,000 base pairs between reads in a pair. For those reads passing the pairing filter, we collected only the read pairs that uniquely mapped as a pair with fewer than five mismatches on each read but did not map anywhere else as a pair with fewer than 12 mismatches on each read to eliminate ambiguous mappings to repetitive genomic regions and false mapping results due to sequencing errors, genetic variations, or RNA editing sites. -

Identification and Functional Annotation of Genes
animals Article Identification and Functional Annotation of Genes Related to Horses’ Performance: From GWAS to Post-GWAS Thayssa O. Littiere 1 , Gustavo H. F. Castro 1, Maria del Pilar R. Rodriguez 1 , Cristina M. Bonafé 1, Ana F. B. Magalhães 1, Rafael R. Faleiros 2, João I. G. Vieira 1, Cassiane G. Santos 1 and Lucas L. Verardo 1,* 1 Department of Animal Science, Universidade Federal dos Vales do Jequitinhonha e Mucuri, Diamantina 39100-000, Brazil; [email protected] (T.O.L.); [email protected] (G.H.F.C.); [email protected] (M.d.P.R.R.); [email protected] (C.M.B.); [email protected] (A.F.B.M.); [email protected] (J.I.G.V.); [email protected] (C.G.S.) 2 EQUINOVA Research Group, Universidade Federal de Minas Gerais, Belo Horizonte 31270-901, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-38-3532-8514 Received: 15 May 2020; Accepted: 8 July 2020; Published: 10 July 2020 Simple Summary: It is assumed that The athletic performance of horses is influenced by a large number of genes; however, to date, not many genomic studies have been performed to identify candidate genes. In this study we performed a systematic review of genome-wide association studies followed by functional analyses aiming to identify The most candidate genes for horse performance. We were successful in identifying 669 candidate genes, from which we built biological process networks. Regulatory elements (transcription factors, TFs) of these genes were identified and used to build a gene–TF network. -
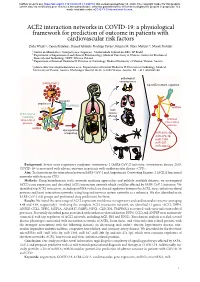
ACE2 Interaction Networks in COVID-19
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.13.094714; this version posted May 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors Zofia Wicik1,2, Ceren Eyileten2, Daniel Jakubik2, Rodrigo Pavão1, Jolanta M. Siller-Matula2,3*, Marek Postula2 ¹ Centro de Matemática, Computação e Cognição - Universidade Federal do ABC, SP, Brazil ² Department of Experimental and Clinical Pharmacology, Medical University of Warsaw, Center for Preclinical Research and Technology CEPT, Warsaw, Poland ³ Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Vienna, Austria * [email protected]; Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Austria. Waehringer Guertel 18-20, A-1090 Vienna, Austria. Tel. +43 1 4040046140 pathological consequences SARS-CoV-2 top ACE2 network regulators activation attachment TMPRSS2 miRNA ACE2 main ACE2 network tissues containing affected virus-related activated proteins virus-related proteins miRNA dysregulation of signaling miR-302c-5p miR-27a-3p miR-1305 miR-587 miR-26b-5p Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcome in patients with cardiovascular disease (CVD). Aim: To characterize the interaction between SARS-CoV-2 and Angiotensin Converting Enzyme 2 (ACE2) functional networks with focus on CVD. -

Integrative Analysis of the Plasma Proteome and Polygenic Risk of Cardiometabolic Diseases
bioRxiv preprint doi: https://doi.org/10.1101/2019.12.14.876474; this version posted December 19, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Integrative analysis of the plasma proteome and polygenic risk of cardiometabolic diseases Scott C. Ritchie1-5,*, Yingying Liu6,7, Samuel A. Lambert1-3,8, Shu Mei Teo1,2, Petar Scepanovic1-3, Jonathan Marten1,3,5, Sohail Zahid9,10, Mark Chaffin9, Gad Abraham1,2,11, Nicole Soranzo12, Stephen Burgess13, Brian G. Drew7,14, Sekar Kathiresan15, Anna C. Calkin6,14, Amit V. Khera9,10,16,17, John Danesh3-5,8,12,18, Adam S. Butterworth3-5,8,12,18, Michael Inouye1-5,8,11,19,* 1Cambridge Baker Systems Genomics Initiative, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK 2Cambridge Baker Systems Genomics Initiative, Baker Heart & Diabetes Institute, Melbourne, Victoria, Australia 3British Heart Foundation Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK 4British Heart Foundation Centre of Research Excellence, University of Cambridge, Cambridge, UK 5National Institute for Health Research Cambridge Biomedical Research Centre, University of Cambridge and Cambridge University Hospitals, Cambridge, UK 6Lipid Metabolism & Cardiometabolic Disease Laboratory, Baker Heart & Diabetes Institute, Melbourne, Victoria, Australia -

Exosomes Derived from Heat Stroke Cases Carry Mirnas Associated with Infammation and Coagulation Cascade
Exosomes Derived From Heat Stroke Cases Carry miRNAs Associated With Inammation and Coagulation Cascade Yue Li Department of ICU, General hospital of southern theatre of PLA Qiang Wen PLA General Hospital of Southern Theatre Command: People's Liberation Army General Hospital of Southern Theatre Command Huaisheng Chen Shenzhen People's Hospital Xinhui Wu PLA General Hospital of Southern Theatre Command: People's Liberation Army General Hospital of Southern Theatre Command Bin Liu third changsha people's hospital Hui Li PLA General Hospital of Southern Theatre Command: People's Liberation Army General Hospital of Southern Theatre Command Lei Su PLA General Hospital of Southern Theatre Command: People's Liberation Army General Hospital of Southern Theatre Command Huasheng Tong ( [email protected] ) Department of Intensive Care Unit, General Hospital of Southern Theatre Command of PLA, 510010, Guangzhou, Guangdong, China. Research Keywords: Heat stroke, Exosome, miRNA, Next generation sequencing Posted Date: October 19th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-92707/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/62 Abstract Background: The pathological mechanism of HS is associated with the dysbalanced inammation and coagulation cascade. The cells-derived circulating extracellular vesicles (EVs) as a novel pathway mediating intercellular communication were evidenced to be associated with immune response and inammation in critical inammatory syndromes such as sepsis. Despite previous studies demonstrating that these vesicles contain genetic material related to their biological function, their molecular cargo during heat stroke is relatively unknown. In this study, we evaluated the presence of microRNAs (miRNAs) and messenger RNAs (mRNAs) related to inammatory response and coagulation cascade in exosomes of patients with heat stroke.