Complete Genome Sequence of Ferroglobus Placidus AEDII12DO
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide
microorganisms Article Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide Kelly J. Hidalgo 1,2,* , Isabel N. Sierra-Garcia 3 , German Zafra 4 and Valéria M. de Oliveira 1 1 Microbial Resources Division, Research Center for Chemistry, Biology and Agriculture (CPQBA), University of Campinas–UNICAMP, Av. Alexandre Cazellato 999, 13148-218 Paulínia, Brazil; [email protected] 2 Graduate Program in Genetics and Molecular Biology, Institute of Biology, University of Campinas (UNICAMP), Rua Monteiro Lobato 255, Cidade Universitária, 13083-862 Campinas, Brazil 3 Biology Department & CESAM, University of Aveiro, Aveiro, Portugal, Campus de Santiago, Avenida João Jacinto de Magalhães, 3810-193 Aveiro, Portugal; [email protected] 4 Grupo de Investigación en Bioquímica y Microbiología (GIBIM), Escuela de Microbiología, Universidad Industrial de Santander, Cra 27 calle 9, 680002 Bucaramanga, Colombia; [email protected] * Correspondence: [email protected]; Tel.: +55-19981721510 Abstract: Microorganisms inhabiting subsurface petroleum reservoirs are key players in biochemical transformations. The interactions of microbial communities in these environments are highly complex and still poorly understood. This work aimed to assess publicly available metagenomes from oil reservoirs and implement a robust pipeline of genome-resolved metagenomics to decipher metabolic and taxonomic profiles of petroleum reservoirs worldwide. Analysis of 301.2 Gb of metagenomic information derived from heavily flooded petroleum reservoirs in China and Alaska to non-flooded petroleum reservoirs in Brazil enabled us to reconstruct 148 metagenome-assembled genomes (MAGs) of high and medium quality. At the phylum level, 74% of MAGs belonged to bacteria and 26% to archaea. The profiles of these MAGs were related to the physicochemical parameters and recovery management applied. -

Marsarchaeota Are an Aerobic Archaeal Lineage Abundant in Geothermal Iron Oxide Microbial Mats
Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats Authors: Zackary J. Jay, Jacob P. Beam, Mansur Dlakic, Douglas B. Rusch, Mark A. Kozubal, and William P. Inskeep This is a postprint of an article that originally appeared in Nature Microbiology on May 14, 2018. The final version can be found at https://dx.doi.org/10.1038/s41564-018-0163-1. Jay, Zackary J. , Jacob P. Beam, Mensur Dlakic, Douglas B. Rusch, Mark A. Kozubal, and William P. Inskeep. "Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats." Nature Microbiology 3, no. 6 (May 2018): 732-740. DOI: 10.1038/ s41564-018-0163-1. Made available through Montana State University’s ScholarWorks scholarworks.montana.edu Marsarchaeota are an aerobic archaeal lineage abundant in geothermal iron oxide microbial mats Zackary J. Jay1,4,7, Jacob P. Beam1,5,7, Mensur Dlakić2, Douglas B. Rusch3, Mark A. Kozubal1,6 and William P. Inskeep 1* The discovery of archaeal lineages is critical to our understanding of the universal tree of life and evolutionary history of the Earth. Geochemically diverse thermal environments in Yellowstone National Park provide unprecedented opportunities for studying archaea in habitats that may represent analogues of early Earth. Here, we report the discovery and character- ization of a phylum-level archaeal lineage proposed and herein referred to as the ‘Marsarchaeota’, after the red planet. The Marsarchaeota contains at least two major subgroups prevalent in acidic, microaerobic geothermal Fe(III) oxide microbial mats across a temperature range from ~50–80 °C. Metagenomics, single-cell sequencing, enrichment culturing and in situ transcrip- tional analyses reveal their biogeochemical role as facultative aerobic chemoorganotrophs that may also mediate the reduction of Fe(III). -

A Web Tool for Hydrogenase Classification and Analysis Dan Søndergaard1, Christian N
www.nature.com/scientificreports OPEN HydDB: A web tool for hydrogenase classification and analysis Dan Søndergaard1, Christian N. S. Pedersen1 & Chris Greening2,3 H2 metabolism is proposed to be the most ancient and diverse mechanism of energy-conservation. The Received: 24 June 2016 metalloenzymes mediating this metabolism, hydrogenases, are encoded by over 60 microbial phyla Accepted: 09 September 2016 and are present in all major ecosystems. We developed a classification system and web tool, HydDB, Published: 27 September 2016 for the structural and functional analysis of these enzymes. We show that hydrogenase function can be predicted by primary sequence alone using an expanded classification scheme (comprising 29 [NiFe], 8 [FeFe], and 1 [Fe] hydrogenase classes) that defines 11 new classes with distinct biological functions. Using this scheme, we built a web tool that rapidly and reliably classifies hydrogenase primary sequences using a combination of k-nearest neighbors’ algorithms and CDD referencing. Demonstrating its capacity, the tool reliably predicted hydrogenase content and function in 12 newly-sequenced bacteria, archaea, and eukaryotes. HydDB provides the capacity to browse the amino acid sequences of 3248 annotated hydrogenase catalytic subunits and also contains a detailed repository of physiological, biochemical, and structural information about the 38 hydrogenase classes defined here. The database and classifier are freely and publicly available at http://services.birc.au.dk/hyddb/ Microorganisms conserve energy by metabolizing H2. Oxidation of this high-energy fuel yields electrons that can be used for respiration and carbon-fixation. This diffusible gas is also produced in diverse fermentation and 1 anaerobic respiratory processes . H2 metabolism contributes to the growth and survival of microorganisms across the three domains of life, including chemotrophs and phototrophs, lithotrophs and heterotrophs, aerobes and 1,2 anaerobes, mesophiles and extremophiles alike . -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Archaeoglobus Profundus Type Strain (AV18T)
Standards in Genomic Sciences (2010) 2:327-346 DOI:10.4056/sigs.942153 Complete genome sequence of Archaeoglobus profundus type strain (AV18T) Mathias von Jan1, Alla Lapidus2, Tijana Glavina Del Rio2, Alex Copeland2, Hope Tice2, Jan-Fang Cheng2, Susan Lucas2, Feng Chen2, Matt Nolan2, Lynne Goodwin2,3, Cliff Han2,3, Sam Pitluck2, Konstantinos Liolios2, Natalia Ivanova2, Konstantinos Mavromatis2, Galina Ovchinnikova2, Olga Chertkov2, Amrita Pati2, Amy Chen4, Krishna Palaniappan4, Miriam Land2,5, Loren Hauser2,5, Yun-Juan Chang2,5, Cynthia D. Jeffries2,5, Elizabeth Saunders2, Thomas Brettin2,3, John C. Detter2,3, Patrick Chain2,4, Konrad Eichinger6, Harald Huber6, Ste- fan Spring1, Manfred Rohde7, Markus Göker1, Reinhard Wirth6, Tanja Woyke2, Jim Bristow2, Jonathan A. Eisen2,8, Victor Markowitz4, Philip Hugenholtz2, Nikos C Kyrpides2, and Hans-Peter Klenk1* 1 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 2 DOE Joint Genome Institute, Walnut Creek, California, USA 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 4 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, USA 5 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 6 University of Regensburg, Microbiology – Archaeenzentrum, Regensburg, Germany 7 HZI – Helmholtz Centre for Infection Research, Braunschweig, Germany 8 University of California Davis Genome Center, Davis, California, USA *Corresponding author: Hans-Peter Klenk Keywords: hyperthermophilic, marine, strictly anaerobic, sulfate respiration, hydrogen utili- zation, hydrothermal systems, Archaeoglobaceae, GEBA Archaeoglobus profundus (Burggraf et al. 1990) is a hyperthermophilic archaeon in the eu- ryarchaeal class Archaeoglobi, which is currently represented by the single family Archaeog- lobaceae, containing six validly named species and two strains ascribed to the genus 'Geoglobus' which is taxonomically challenged as the corresponding type species has no va- lidly published name. -

Assessment of the Carbon Monoxide Metabolism of the Hyperthermophilic Sulfate-Reducing Archaeon Archaeoglobus Fulgidus VC-16 by Comparative Transcriptome Analyses
Hindawi Publishing Corporation Archaea Volume 2015, Article ID 235384, 12 pages http://dx.doi.org/10.1155/2015/235384 Research Article Assessment of the Carbon Monoxide Metabolism of the Hyperthermophilic Sulfate-Reducing Archaeon Archaeoglobus fulgidus VC-16 by Comparative Transcriptome Analyses William P. Hocking, Irene Roalkvam, Carina Magnussen, Runar Stokke, and Ida H. Steen Department of Biology, Centre for Geobiology, University of Bergen, 5020 Bergen, Norway Correspondence should be addressed to Ida H. Steen; [email protected] Received 10 April 2015; Revised 9 June 2015; Accepted 14 June 2015 Academic Editor: Uwe Deppenmeier Copyright © 2015 William P. Hocking et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The hyperthermophilic, sulfate-reducing archaeon, Archaeoglobus fulgidus, utilizes CO as an energy source and it is resistant to the toxic effects of high CO concentrations. Herein, transcription profiles were obtained from A. fulgidus during growth with CO and sulfate or thiosulfate, or without an electron acceptor. This provided a basis for a model of the CO metabolism of A. fulgidus. The model suggests proton translocation by “Mitchell-type” loops facilitated by Fqo catalyzingred aFd :menaquinone oxidoreductase reaction, as the major mode of energy conservation, rather than formate or H2 cycling during respiratory growth. The bifunctional CODH (cdhAB-2) is predicted to play an ubiquitous role in the metabolism of CO, and a novel nitrate reductase- associated respiratory complex was induced specifically in the presence of sulfate. A potential role of this complex in relation to Fdred and APS reduction is discussed. -

Specific Enrichment of Hyperthermophilic Electroactive Archaea From
bioRxiv preprint doi: https://doi.org/10.1101/272039; this version posted February 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Specific enrichment of hyperthermophilic electroactive Archaea from 2 deep-sea hydrothermal vent on electrically conductive support. 3 4 Guillaume Pillot1, Eléonore Frouin1, Emilie Pasero1, Anne Godfroy2, Yannick Combet-Blanc1, 5 Sylvain Davidson1 & Pierre-Pol Liebgott1* 6 1 Aix Marseille Université, IRD, Université de Toulon, CNRS, MIO UM 110, Marseille, France. 7 2 Laboratoire de Microbiologie des Environnements Extrêmes - UMR6197 IFREMER, CNRS, 8 UBO Centre de Brest–CS10070/IUEM, Plouzané, France. 9 *Corresponding author: [email protected]; Mediterranean Institute of 10 Oceanography, Campus de Luminy, Bâtiment OCEANOMED, 13288 Marseille Cedex 09. 11 Abstract 12 While more and more investigations are done to isolate hyperthermophilic exoelectrogenic 13 communities from environments, none have been performed yet on deep-sea hydrothermal vent. 14 Samples of black smoker chimney from Rainbow site on the Atlantic mid-oceanic ridge have 15 been harvested for enriching exoelectrogens in microbial electrolysis cells under hyperthermophilic 16 (80°C) condition. Two enrichments have been performed: one from direct inoculation of crushed 17 chimney and the other one from inoculation of a pre-cultivation on iron (III) oxide. In both 18 experiments, a current production was observed from 2.4 A/m² to 5.8 A/m² with a set anode 19 potential of +0.05 vs SHE. Taxonomic affiliation of the exoelectrogen communities obtained 20 exhibited a specific enrichment of Archaea from Thermococcales and Archeoglobales orders on the 21 electrode, even when both inocula were dominated by Bacteria. -

Thermophilic Carboxydotrophs and Their Applications in Biotechnology Springerbriefs in Microbiology
SPRINGER BRIEFS IN MICROBIOLOGY EXTREMOPHILIC BACTERIA Sonia M. Tiquia-Arashiro Thermophilic Carboxydotrophs and their Applications in Biotechnology SpringerBriefs in Microbiology Extremophilic Bacteria Series editors Sonia M. Tiquia-Arashiro, Dearborn, MI, USA Melanie Mormile, Rolla, MO, USA More information about this series at http://www.springer.com/series/11917 Sonia M. Tiquia-Arashiro Thermophilic Carboxydotrophs and their Applications in Biotechnology 123 Sonia M. Tiquia-Arashiro Department of Natural Sciences University of Michigan Dearborn, MI USA ISSN 2191-5385 ISSN 2191-5393 (electronic) ISBN 978-3-319-11872-7 ISBN 978-3-319-11873-4 (eBook) DOI 10.1007/978-3-319-11873-4 Library of Congress Control Number: 2014951696 Springer Cham Heidelberg New York Dordrecht London © The Author(s) 2014 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. Exempted from this legal reservation are brief excerpts in connection with reviews or scholarly analysis or material supplied specifically for the purpose of being entered and executed on a computer system, for exclusive use by the purchaser of the work. Duplication of this publication or parts thereof is permitted only under the provisions of the Copyright Law of the Publisher’s location, in its current version, and permission for use must always be obtained from Springer. -

Deep-Sea Hydrothermal Vent Euryarchaeota 2”
View metadata, citation and similar papers at core.ac.uk brought to you by CORE ORIGINAL RESEARCH ARTICLE published: 20 February 2012provided by PubMed Central doi: 10.3389/fmicb.2012.00047 Distribution, abundance, and diversity patterns of the thermoacidophilic “deep-sea hydrothermal vent euryarchaeota 2” Gilberto E. Flores†, Isaac D. Wagner,Yitai Liu and Anna-Louise Reysenbach* Department of Biology, Center for Life in Extreme Environments, Portland State University, Portland, OR, USA Edited by: Cultivation-independent studies have shown that taxa belonging to the “deep-sea Kirsten Silvia Habicht, University of hydrothermal vent euryarchaeota 2” (DHVE2) lineage are widespread at deep-sea Southern Denmark, Denmark hydrothermal vents. While this lineage appears to be a common and important mem- Reviewed by: Kuk-Jeong Chin, Georgia State ber of the microbial community at vent environments, relatively little is known about their University, USA overall distribution and phylogenetic diversity. In this study, we examined the distribu- Elizaveta Bonch-Osmolovskyaya, tion, relative abundance, co-occurrence patterns, and phylogenetic diversity of cultivable Winogradsky Institute of Microbiology thermoacidophilic DHVE2 in deposits from globally distributed vent fields. Results of quan- Russian Academy of Sciences, Russia titative polymerase chain reaction assays with primers specific for the DHVE2 and Archaea *Correspondence: Anna-Louise Reysenbach, demonstrate the ubiquity of the DHVE2 at deep-sea vents and suggest that they are sig- Department of Biology, Center for nificant members of the archaeal communities of established vent deposit communities. Life in Extreme Environments, Local similarity analysis of pyrosequencing data revealed that the distribution of the DHVE2 Portland State University, PO Box was positively correlated with 10 other Euryarchaeota phylotypes and negatively correlated 751, Portland, OR 97207-0751, USA. -

Microbial and Geochemical Investigation Down to 2000 M Deep Triassic Rock (Meuse/Haute Marne, France)
geosciences Article Microbial and Geochemical Investigation down to 2000 m Deep Triassic Rock (Meuse/Haute Marne, France) Vanessa Leblanc 1,2,3, Jennifer Hellal 1 , Marie-Laure Fardeau 4,5, Saber Khelaifia 4,5, Claire Sergeant 2,3, Francis Garrido 1, Bernard Ollivier 4,5 and Catherine Joulian 1,* 1 BRGM, Geomicrobiology and Environmental Monitoring Unit, F-45060 Orléans CEDEX 02, France; [email protected] (V.L.); [email protected] (J.H.); [email protected] (F.G.) 2 Bordeaux University, Centre d’Etudes Nucleaires de Bordeaux Gradignan, UMR5797, F-33170 Gradignan, France; [email protected] 3 CNRS-IN2P3, Centre d’Etudes Nucleaires de Bordeaux Gradignan, UMR5797, F-33170 Gradignan, France 4 Aix Marseille Université, CNRS/INSU, IRD, Mediterranean Institute of Oceanography (MIO), UM 110, 13288 Marseille, France; [email protected] (M.-L.F.); Saber.Khelaifi[email protected] (S.K.); [email protected] (B.O.) 5 Université de Toulon, CNRS/INSU, 83957 La Garde, France * Correspondence: [email protected]; Tel.: +33-2-3864-3089 Received: 31 August 2018; Accepted: 13 December 2018; Published: 20 December 2018 Abstract: In 2008, as part of a feasibility study for radioactive waste disposal in deep geological formations, the French National Radioactive Waste Management Agency (ANDRA) drilled several boreholes in the transposition zone in order to define the potential variations in the properties of the Callovo–Oxfordian claystone formation. This consisted of a rare opportunity to investigate the deep continental biosphere that is still poorly known. Four rock cores, from 1709, 1804, 1865, and 1935 m below land surface, were collected from Lower and Middle Triassic formations in the Paris Basin (France) to investigate their microbial and geochemical composition. -

Assessment of the Carbon Monoxide Metabolism of the Hyperthermophilic Sulfate-Reducing Archaeon Archaeoglobus Fulgidus VC-16 by Comparative Transcriptome Analyses
Hindawi Publishing Corporation Archaea Volume 2015, Article ID 235384, 12 pages http://dx.doi.org/10.1155/2015/235384 Research Article Assessment of the Carbon Monoxide Metabolism of the Hyperthermophilic Sulfate-Reducing Archaeon Archaeoglobus fulgidus VC-16 by Comparative Transcriptome Analyses William P. Hocking, Irene Roalkvam, Carina Magnussen, Runar Stokke, and Ida H. Steen Department of Biology, Centre for Geobiology, University of Bergen, 5020 Bergen, Norway Correspondence should be addressed to Ida H. Steen; [email protected] Received 10 April 2015; Revised 9 June 2015; Accepted 14 June 2015 Academic Editor: Uwe Deppenmeier Copyright © 2015 William P. Hocking et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The hyperthermophilic, sulfate-reducing archaeon, Archaeoglobus fulgidus, utilizes CO as an energy source and it is resistant to the toxic effects of high CO concentrations. Herein, transcription profiles were obtained from A. fulgidus during growth with CO and sulfate or thiosulfate, or without an electron acceptor. This provided a basis for a model of the CO metabolism of A. fulgidus. The model suggests proton translocation by “Mitchell-type” loops facilitated by Fqo catalyzingred aFd :menaquinone oxidoreductase reaction, as the major mode of energy conservation, rather than formate or H2 cycling during respiratory growth. The bifunctional CODH (cdhAB-2) is predicted to play an ubiquitous role in the metabolism of CO, and a novel nitrate reductase- associated respiratory complex was induced specifically in the presence of sulfate. A potential role of this complex in relation to Fdred and APS reduction is discussed. -
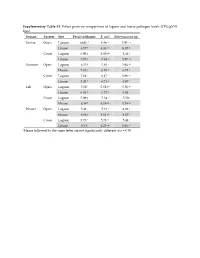
Supplementary Table S1. Fisher Pairwise Comparisons of Lagoon and House Pathogen Levels (CFU/Gvss Log10) Season System Site Fecal Coliforms E
Supplementary Table S1. Fisher pairwise comparisons of lagoon and house pathogen levels (CFU/gVSS log10) Season System Site Fecal coliforms E. coli Enterococcus sp. Spring Open Lagoon 6.00 g1 5.46 hi 5.91 cd House 6.57 e 6.33 cd 6.35 b Cover Lagoon 6.00 g 5.60 gh 5.44 f House 7.73 a 7.63 a 5.87 cd Summer Open Lagoon 6.35 f 5.83 f 5.94 cd House 7.83 a 6.10 e 6.73 a Cover Lagoon 7.04 c 6.47 c 5.86 cd House 7.42 b 6.71 b 6.07 c Fall Open Lagoon 5.58 i 5.58 gh 5.56 ef House 6.81 d 6.72 b 6.01 c Cover Lagoon 5.99 g 5.34 i 5.50 f House 6.38 f 6.19 de 5.74 de Winter Open Lagoon 5.41 j 5.13 j 4.88 g House 6.06 g 5.61 gh 6.67 a Cover Lagoon 5.73 h 5.73 fg 5.44 f House 6.34 f 6.25 de 5.86 cd 1Means followed by the same letter are not significantly different at p = 0.05 Supplementary Table S2. Relative abundances of OTUs identified using universal bacterial primer set, presented as relative abundances (%) Only bacterial families are counted in Figure 5 and discussion involving bacterial family identification. # Identified in # Identified from all 8 All 16 All #OTU Table SpOLRA SuOLRA FOLRA WOLRA SpCLRA SuCLRA FCLRA WCLRA SpOHRA SuOHRA FOHRA WOHRA SpCHRA SuCHRA FCHRA WCHRA all samples lagoon samples Samples Lagoons Notes Key Patulibacteraceae 0 9.93739E‐05* 0.000134 0.000116 0 0 0 0 0 0 0.000452 0 0 0 0 0 4 3 Sp = Spring O = Open Ruaniaceae 0 0 0 0 0 0 0 0 0 0 0 0 0 0.000221577 0 7.73E‐05 2 0 Su = Summer C = Covered Thermoactinomycetaceae 0 0 0.000267 0.000116 0 0.00037092 0.000529 0.000526 0 0.000342922 0.000271 0.000197 0 0 0 0 8 5 F = Fall L = Lagoon Beutenbergiaceae 0.000226334 0.000331246 0.000579 0.001663 0.000620176 0 0.000106 0.000807 0.002320743 0.000571537 0.000723 0.000591 0.000169731 0 0.000606 0 13 7 W = Winter H = House Thermoanaerobacterales Family III.