Systematic Bacteriology Second Edition
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide
microorganisms Article Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide Kelly J. Hidalgo 1,2,* , Isabel N. Sierra-Garcia 3 , German Zafra 4 and Valéria M. de Oliveira 1 1 Microbial Resources Division, Research Center for Chemistry, Biology and Agriculture (CPQBA), University of Campinas–UNICAMP, Av. Alexandre Cazellato 999, 13148-218 Paulínia, Brazil; [email protected] 2 Graduate Program in Genetics and Molecular Biology, Institute of Biology, University of Campinas (UNICAMP), Rua Monteiro Lobato 255, Cidade Universitária, 13083-862 Campinas, Brazil 3 Biology Department & CESAM, University of Aveiro, Aveiro, Portugal, Campus de Santiago, Avenida João Jacinto de Magalhães, 3810-193 Aveiro, Portugal; [email protected] 4 Grupo de Investigación en Bioquímica y Microbiología (GIBIM), Escuela de Microbiología, Universidad Industrial de Santander, Cra 27 calle 9, 680002 Bucaramanga, Colombia; [email protected] * Correspondence: [email protected]; Tel.: +55-19981721510 Abstract: Microorganisms inhabiting subsurface petroleum reservoirs are key players in biochemical transformations. The interactions of microbial communities in these environments are highly complex and still poorly understood. This work aimed to assess publicly available metagenomes from oil reservoirs and implement a robust pipeline of genome-resolved metagenomics to decipher metabolic and taxonomic profiles of petroleum reservoirs worldwide. Analysis of 301.2 Gb of metagenomic information derived from heavily flooded petroleum reservoirs in China and Alaska to non-flooded petroleum reservoirs in Brazil enabled us to reconstruct 148 metagenome-assembled genomes (MAGs) of high and medium quality. At the phylum level, 74% of MAGs belonged to bacteria and 26% to archaea. The profiles of these MAGs were related to the physicochemical parameters and recovery management applied. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Genotyping of Uncultured Archaea in a Polluted Site of Suez Gulf, Egypt, Based on 16S Rrna Gene Analyses
Egyptian Journal of Aquatic Research (2014) 40,27–33 National Institute of Oceanography and Fisheries Egyptian Journal of Aquatic Research http://ees.elsevier.com/ejar www.sciencedirect.com FULL LENGTH ARTICLE Genotyping of uncultured archaea in a polluted site of Suez Gulf, Egypt, based on 16S rRNA gene analyses Hosam Easa Elsaied * Aquagenome Resources and Biotechnology Research Group, National Institute of Oceanography, 101-El-Kasr El-Eini Street, Cairo, Egypt Received 3 February 2014; revised 11 March 2014; accepted 11 March 2014 Available online 18 April 2014 KEYWORDS Abstract Culture-independent 16S rRNA gene analysis approach was used to explore and evalu- Archaea; ate archaea in a polluted site, El-Zeitia, Suez Gulf, Egypt. Metagenomic DNA was extracted from a 16S rRNA gene diversity; sediment sample. Archaeal 16S rRNA gene was PCR amplified using universal archaeal primers, Sediment; followed by cloning and direct analyses by sequencing. Rarefaction analysis showed saturation, Suez Gulf recording 21 archaeal 16S rRNA gene phylotypes, which represented the total composition of archaea in the studied sample. Phylogenetic analysis showed that all recorded phylotypes belonged to two archaeal phyla. Sixteen phylotypes were located in the branch of methanogenic Eur- yarchaeota and more closely related to species of the genera Methanosaeta and Methanomassiliicoc- cus. Five phylotypes were affiliated to the new archaeal phylum Thaumarchaeota, which represented by species Candidatus nitrosopumilus. The recorded phylotypes had unique sequences, characteriz- ing them as new phylogenetic lineages. This work is the first investigation of uncultured archaea in the Suez Gulf, and implicated that the environmental characteristics shaped the diversity of archa- eal 16S rRNA genes in the studied sample. -

Archaeoglobus Profundus Type Strain (AV18T)
Standards in Genomic Sciences (2010) 2:327-346 DOI:10.4056/sigs.942153 Complete genome sequence of Archaeoglobus profundus type strain (AV18T) Mathias von Jan1, Alla Lapidus2, Tijana Glavina Del Rio2, Alex Copeland2, Hope Tice2, Jan-Fang Cheng2, Susan Lucas2, Feng Chen2, Matt Nolan2, Lynne Goodwin2,3, Cliff Han2,3, Sam Pitluck2, Konstantinos Liolios2, Natalia Ivanova2, Konstantinos Mavromatis2, Galina Ovchinnikova2, Olga Chertkov2, Amrita Pati2, Amy Chen4, Krishna Palaniappan4, Miriam Land2,5, Loren Hauser2,5, Yun-Juan Chang2,5, Cynthia D. Jeffries2,5, Elizabeth Saunders2, Thomas Brettin2,3, John C. Detter2,3, Patrick Chain2,4, Konrad Eichinger6, Harald Huber6, Ste- fan Spring1, Manfred Rohde7, Markus Göker1, Reinhard Wirth6, Tanja Woyke2, Jim Bristow2, Jonathan A. Eisen2,8, Victor Markowitz4, Philip Hugenholtz2, Nikos C Kyrpides2, and Hans-Peter Klenk1* 1 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 2 DOE Joint Genome Institute, Walnut Creek, California, USA 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, New Mexico, USA 4 Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, USA 5 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 6 University of Regensburg, Microbiology – Archaeenzentrum, Regensburg, Germany 7 HZI – Helmholtz Centre for Infection Research, Braunschweig, Germany 8 University of California Davis Genome Center, Davis, California, USA *Corresponding author: Hans-Peter Klenk Keywords: hyperthermophilic, marine, strictly anaerobic, sulfate respiration, hydrogen utili- zation, hydrothermal systems, Archaeoglobaceae, GEBA Archaeoglobus profundus (Burggraf et al. 1990) is a hyperthermophilic archaeon in the eu- ryarchaeal class Archaeoglobi, which is currently represented by the single family Archaeog- lobaceae, containing six validly named species and two strains ascribed to the genus 'Geoglobus' which is taxonomically challenged as the corresponding type species has no va- lidly published name. -

Research Article Diversity and Distribution of Archaea in the Mangrove Sediment of Sundarbans
Hindawi Publishing Corporation Archaea Volume 2015, Article ID 968582, 14 pages http://dx.doi.org/10.1155/2015/968582 Research Article Diversity and Distribution of Archaea in the Mangrove Sediment of Sundarbans Anish Bhattacharyya,1 Niladri Shekhar Majumder,2 Pijush Basak,1 Shayantan Mukherji,3 Debojyoti Roy,1 Sudip Nag,1 Anwesha Haldar,4 Dhrubajyoti Chattopadhyay,1 Suparna Mitra,5 Maitree Bhattacharyya,1 and Abhrajyoti Ghosh3 1 Department of Biochemistry, University of Calcutta, 35 Ballygunge Circular Road, Kolkata, West Bengal 700019, India 2RocheDiagnosticsIndiaPvt.Ltd.,Block4C,AkashTower,NearRubyHospital,781Anandapur,Kolkata700107,India 3Department of Biochemistry, Bose Institute, P1/12, C. I. T. Road, Scheme VIIM, Kolkata, West Bengal 700054, India 4Department of Geography, University of Calcutta, 35 Ballygunge Circular Road, Kolkata, West Bengal 700019, India 5Norwich Medical School, University of East Anglia and Institute of Food Research, Norwich Research Park, Norwich, Norfolk NR4 7UA, UK Correspondence should be addressed to Dhrubajyoti Chattopadhyay; [email protected], Suparna Mitra; [email protected], Maitree Bhattacharyya; [email protected], and Abhrajyoti Ghosh; [email protected] Received 30 March 2015; Revised 25 June 2015; Accepted 14 July 2015 Academic Editor: William B. Whitman Copyright © 2015 Anish Bhattacharyya et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Mangroves are among the most diverse and productive coastal ecosystems in the tropical and subtropical regions. Environmental conditions particular to this biome make mangroves hotspots for microbial diversity, and the resident microbial communities play essential roles in maintenance of the ecosystem. -

Supporting Information
Supporting Information Lozupone et al. 10.1073/pnas.0807339105 SI Methods nococcus, and Eubacterium grouped with members of other Determining the Environmental Distribution of Sequenced Genomes. named genera with high bootstrap support (Fig. 1A). One To obtain information on the lifestyle of the isolate and its reported member of the Bacteroidetes (Bacteroides capillosus) source, we looked at descriptive information from NCBI grouped firmly within the Firmicutes. This taxonomic error was (www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and other related not surprising because gut isolates have often been classified as publications. We also determined which 16S rRNA-based envi- Bacteroides based on an obligate anaerobe, Gram-negative, ronmental surveys of microbial assemblages deposited near- nonsporulating phenotype alone (6, 7). A more recent 16S identical sequences in GenBank. We first downloaded the gbenv rRNA-based analysis of the genus Clostridium defined phylo- files from the NCBI ftp site on December 31, 2007, and used genetically related clusters (4, 5), and these designations were them to create a BLAST database. These files contain GenBank supported in our phylogenetic analysis of the Clostridium species in the HGMI pipeline. We thus designated these Clostridium records for the ENV database, a component of the nonredun- species, along with the species from other named genera that dant nucleotide database (nt) where 16S rRNA environmental cluster with them in bootstrap supported nodes, as being within survey data are deposited. GenBank records for hits with Ͼ98% these clusters. sequence identity over 400 bp to the 16S rRNA sequence of each of the 67 genomes were parsed to get a list of study titles Annotation of GTs and GHs. -

Insights Into Archaeal Evolution and Symbiosis from the Genomes of a Nanoarchaeon and Its Inferred Crenarchaeal Host from Obsidian Pool, Yellowstone National Park
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Microbiology Publications and Other Works Microbiology 4-22-2013 Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar University of Tennessee - Knoxville, [email protected] Kira S. Makarova National Institutes of Health David E. Graham University of Tennessee - Knoxville, [email protected] Yuri I. Wolf National Institutes of Health Eugene V. Koonin National Institutes of Health See next page for additional authors Follow this and additional works at: https://trace.tennessee.edu/utk_micrpubs Part of the Microbiology Commons Recommended Citation Biology Direct 2013, 8:9 doi:10.1186/1745-6150-8-9 This Article is brought to you for free and open access by the Microbiology at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Microbiology Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Authors Mircea Podar, Kira S. Makarova, David E. Graham, Yuri I. Wolf, Eugene V. Koonin, and Anna-Louise Reysenbach This article is available at TRACE: Tennessee Research and Creative Exchange: https://trace.tennessee.edu/ utk_micrpubs/44 Podar et al. Biology Direct 2013, 8:9 http://www.biology-direct.com/content/8/1/9 RESEARCH Open Access Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar1,2*, Kira S Makarova3, David E Graham1,2, Yuri I Wolf3, Eugene V Koonin3 and Anna-Louise Reysenbach4 Abstract Background: A single cultured marine organism, Nanoarchaeum equitans, represents the Nanoarchaeota branch of symbiotic Archaea, with a highly reduced genome and unusual features such as multiple split genes. -
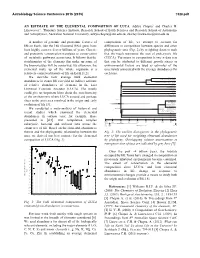
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Brian W. Waters
INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. WATERS (Under the direction of Dr. William B. Whitman) ABSTRACT With the emergence of Methanococcus maripaludis as a genetic model for methanogens, it becomes imperative to devise inexpensive yet effective ways to grow large numbers of cells for protein studies. Chapter 2 details methods that were used to cut costs of growing M. maripaludis in large scale. Also, large scale growth under different conditions was explored in order to find the conditions that yield the most cells. Chapter 3 details the phylogenetic analysis of 2-oxoacid oxidoreductase (OR) homologs from M. maripaludis. ORs are enzymes that catalyze the oxidative decarboxylation of 2-oxoacids to their acyl-CoA derivatives in many prokaryotes. Also in chapter 3, a specific OR homolog in M. maripaludis, an indolepyruvate oxidoreductase (IOR), was mutagenized, and the mutant was characterized. PCR and Southern hybridization analysis showed gene replacement. A no-growth phenotype on media containing aromatic amino acid derivatives was found. INDEX WORDS: Methanococcus maripaludis, fermentor, 2-oxoacid oxidoreductase, indolepyruvate oxidoreductase INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. WATERS B.S., The University of Georgia,1999 A Thesis Submitted to the Graduate Faculty of The University of Georgia in Partial Fulfillment of the Requirements for the Degree MASTER OF SCIENCE ATHENS, GEORGIA 2002 © 2002 Brian W. Waters All Rights Reserved INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. -

Pan-Genome Analysis and Ancestral State Reconstruction Of
www.nature.com/scientificreports OPEN Pan‑genome analysis and ancestral state reconstruction of class halobacteria: probability of a new super‑order Sonam Gaba1,2, Abha Kumari2, Marnix Medema 3 & Rajeev Kaushik1* Halobacteria, a class of Euryarchaeota are extremely halophilic archaea that can adapt to a wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl. It consists of the orders: Halobacteriales, Haloferaciales and Natriabales. Pan‑genome analysis of class Halobacteria was done to explore the core (300) and variable components (Softcore: 998, Cloud:36531, Shell:11784). The core component revealed genes of replication, transcription, translation and repair, whereas the variable component had a major portion of environmental information processing. The pan‑gene matrix was mapped onto the core‑gene tree to fnd the ancestral (44.8%) and derived genes (55.1%) of the Last Common Ancestor of Halobacteria. A High percentage of derived genes along with presence of transformation and conjugation genes indicate the occurrence of horizontal gene transfer during the evolution of Halobacteria. A Core and pan‑gene tree were also constructed to infer a phylogeny which implicated on the new super‑order comprising of Natrialbales and Halobacteriales. Halobacteria1,2 is a class of phylum Euryarchaeota3 consisting of extremely halophilic archaea found till date and contains three orders namely Halobacteriales4,5 Haloferacales5 and Natrialbales5. Tese microorganisms are able to dwell at wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl6. Halobacteria, as the name suggests were once considered a part of a domain "Bacteria" but with the discovery of the third domain "Archaea" by Carl Woese et al.7, it became part of Archaea. -

(Gid ) Genes Coding for Putative Trna:M5u-54 Methyltransferases in 355 Bacterial and Archaeal Complete Genomes
Table S1. Taxonomic distribution of the trmA and trmFO (gid ) genes coding for putative tRNA:m5U-54 methyltransferases in 355 bacterial and archaeal complete genomes. Asterisks indicate the presence and the number of putative genes found. Genomes Taxonomic position TrmA Gid Archaea Crenarchaea Aeropyrum pernix_K1 Crenarchaeota; Thermoprotei; Desulfurococcales; Desulfurococcaceae Cenarchaeum symbiosum Crenarchaeota; Thermoprotei; Cenarchaeales; Cenarchaeaceae Pyrobaculum aerophilum_str_IM2 Crenarchaeota; Thermoprotei; Thermoproteales; Thermoproteaceae Sulfolobus acidocaldarius_DSM_639 Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae Sulfolobus solfataricus Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae Sulfolobus tokodaii Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae Euryarchaea Archaeoglobus fulgidus Euryarchaeota; Archaeoglobi; Archaeoglobales; Archaeoglobaceae Haloarcula marismortui_ATCC_43049 Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Haloarcula Halobacterium sp Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Haloarcula Haloquadratum walsbyi Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Haloquadra Methanobacterium thermoautotrophicum Euryarchaeota; Methanobacteria; Methanobacteriales; Methanobacteriaceae Methanococcoides burtonii_DSM_6242 Euryarchaeota; Methanomicrobia; Methanosarcinales; Methanosarcinaceae Methanococcus jannaschii Euryarchaeota; Methanococci; Methanococcales; Methanococcaceae Methanococcus maripaludis_S2 Euryarchaeota; Methanococci;