Shigella - Shigellosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Shigella Infection - Factsheet
Shigella Infection - Factsheet What is Shigellosis? How common is it? Shigellosis is an infectious disease caused by a group of bacteria (germs) called Shigella. It’s also known as bacillary dysentery. There are four main types of Shigella germ but Shigella sonnei is by far the commonest cause of this illness in the UK. Most cases of the other types are usually brought in from abroad. How is Shigellosis caught? Shigella is not known to be found in animals so it always passes from one infected person to the next, though the route may be indirect. Here are some possible ways in which you can get infected: • Shigella germs are present in the stools of infected persons while they are ill and for a week or two afterwards. Most Shigella infections are the result of germs passing from stools or soiled fingers of one person to the mouth of another person. This happens when basic hygiene and hand washing habits are inadequate, such as in young toddlers who are not yet fully toilet trained. Family members and playmates of such children are at high risk of becoming infected. • Shigellosis can be acquired from someone who is infected but has no symptoms. • Shigellosis may be picked up from eating contaminated food, which may look and smell normal. Food may become contaminated by infected food handlers who do not wash their hands properly after using the toilet. They should report sick and avoid handling food if they are ill but they may not always have symptoms. • Vegetables can become contaminated if they are harvested from a field with sewage in it. -

Succession and Persistence of Microbial Communities and Antimicrobial Resistance Genes Associated with International Space Stati
Singh et al. Microbiome (2018) 6:204 https://doi.org/10.1186/s40168-018-0585-2 RESEARCH Open Access Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces Nitin Kumar Singh1, Jason M. Wood1, Fathi Karouia2,3 and Kasthuri Venkateswaran1* Abstract Background: The International Space Station (ISS) is an ideal test bed for studying the effects of microbial persistence and succession on a closed system during long space flight. Culture-based analyses, targeted gene-based amplicon sequencing (bacteriome, mycobiome, and resistome), and shotgun metagenomics approaches have previously been performed on ISS environmental sample sets using whole genome amplification (WGA). However, this is the first study reporting on the metagenomes sampled from ISS environmental surfaces without the use of WGA. Metagenome sequences generated from eight defined ISS environmental locations in three consecutive flights were analyzed to assess the succession and persistence of microbial communities, their antimicrobial resistance (AMR) profiles, and virulence properties. Metagenomic sequences were produced from the samples treated with propidium monoazide (PMA) to measure intact microorganisms. Results: The intact microbial communities detected in Flight 1 and Flight 2 samples were significantly more similar to each other than to Flight 3 samples. Among 318 microbial species detected, 46 species constituting 18 genera were common in all flight samples. Risk group or biosafety level 2 microorganisms that persisted among all three flights were Acinetobacter baumannii, Haemophilus influenzae, Klebsiella pneumoniae, Salmonella enterica, Shigella sonnei, Staphylococcus aureus, Yersinia frederiksenii,andAspergillus lentulus.EventhoughRhodotorula and Pantoea dominated the ISS microbiome, Pantoea exhibited succession and persistence. K. pneumoniae persisted in one location (US Node 1) of all three flights and might have spread to six out of the eight locations sampled on Flight 3. -

Recognizing and Treating New and Emerging Infections Encountered in Everyday Practice
Recognizing and treating new and emerging infections encountered in everyday practice STEVEN M. GORDON, MD NFECTIOUS DISEASES, pre- MiikWirj:« Although infectious diseases were once considered a dicted earlier in this cen- diminishing threat, new pathogens are constantly challenging tury to be eliminated as a the health care system. This article reviews the clinical presen- public health problem, re- tation, diagnosis, and treatment of seven emerging infections I main the chief cause of death that primary care physicians are likely to encounter. worldwide and a significant cause of death and morbidity in i Parvovirus B19 attacks erythrocyte precursors; the United States.1 Challenging infection is usually benign and self-limiting but can cause the US public health system are aplastic crises in patients with chronic hemolytic disorders. several newly identified patho- Hemorrhagic colitis due to Escherichia coli 0157:H7 infection gens (eg, human immunodefi- can lead to the hemolytic-uremic syndrome, especially in chil- ciency virus [HIV], Escherichia dren; it also can cause thrombotic thrombocytopenia purpura. coli 0157:H7, hepatitis C) and a Chlamydia pneumoniae causes a mild pneumonia that resem- resurgence of old diseases pre- bles mycoplasmal pneumonia. Bacillary angiomatosis primar- sumed to be under control (eg, ily affects immunocompromised patients, especially those tuberculosis, syphilis). Further, infected with human immunodeficiency virus (HIV). At least multiple-drug resistance in two organisms can cause bacillary angiomatosis: Bartonella hense- strains of pneumococci, gono- lae and Bartonella quintana. Hantavirus pulmonary syndrome cocci, enterococci, staphylo- is spread by exposure to the droppings of infected rodents. cocci, salmonella, and mycobac- Contrary to previous thought, HIV continues to replicate teria undermines efforts to throughout the course of the illness and does not have a latency control the diseases they cause.2 phase. -

Bacterial Communities of the Upper Respiratory Tract of Turkeys
www.nature.com/scientificreports OPEN Bacterial communities of the upper respiratory tract of turkeys Olimpia Kursa1*, Grzegorz Tomczyk1, Anna Sawicka‑Durkalec1, Aleksandra Giza2 & Magdalena Słomiany‑Szwarc2 The respiratory tracts of turkeys play important roles in the overall health and performance of the birds. Understanding the bacterial communities present in the respiratory tracts of turkeys can be helpful to better understand the interactions between commensal or symbiotic microorganisms and other pathogenic bacteria or viral infections. The aim of this study was the characterization of the bacterial communities of upper respiratory tracks in commercial turkeys using NGS sequencing by the amplifcation of 16S rRNA gene with primers designed for hypervariable regions V3 and V4 (MiSeq, Illumina). From 10 phyla identifed in upper respiratory tract in turkeys, the most dominated phyla were Firmicutes and Proteobacteria. Diferences in composition of bacterial diversity were found at the family and genus level. At the genus level, the turkey sequences present in respiratory tract represent 144 established bacteria. Several respiratory pathogens that contribute to the development of infections in the respiratory system of birds were identifed, including the presence of Ornithobacterium and Mycoplasma OTUs. These results obtained in this study supply information about bacterial composition and diversity of the turkey upper respiratory tract. Knowledge about bacteria present in the respiratory tract and the roles they can play in infections can be useful in controlling, diagnosing and treating commercial turkey focks. Next-generation sequencing has resulted in a marked increase in culture-independent studies characterizing the microbiome of humans and animals1–6. Much of these works have been focused on the gut microbiome of humans and other production animals 7–11. -
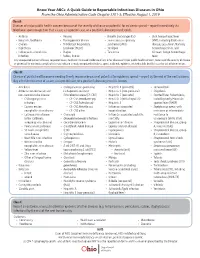
Know Your Abcs: a Quick Guide to Reportable Infectious Diseases in Ohio
Know Your ABCs: A Quick Guide to Reportable Infectious Diseases in Ohio From the Ohio Administrative Code Chapter 3701-3; Effective August 1, 2019 Class A: Diseases of major public health concern because of the severity of disease or potential for epidemic spread – report immediately via telephone upon recognition that a case, a suspected case, or a positive laboratory result exists. • Anthrax • Measles • Rubella (not congenital) • Viral hemorrhagic fever • Botulism, foodborne • Meningococcal disease • Severe acute respiratory (VHF), including Ebola virus • Cholera • Middle East Respiratory syndrome (SARS) disease, Lassa fever, Marburg • Diphtheria Syndrome (MERS) • Smallpox hemorrhagic fever, and • Influenza A – novel virus • Plague • Tularemia Crimean-Congo hemorrhagic infection • Rabies, human fever Any unexpected pattern of cases, suspected cases, deaths or increased incidence of any other disease of major public health concern, because of the severity of disease or potential for epidemic spread, which may indicate a newly recognized infectious agent, outbreak, epidemic, related public health hazard or act of bioterrorism. Class B: Disease of public health concern needing timely response because of potential for epidemic spread – report by the end of the next business day after the existence of a case, a suspected case, or a positive laboratory result is known. • Amebiasis • Carbapenemase-producing • Hepatitis B (perinatal) • Salmonellosis • Arboviral neuroinvasive and carbapenem-resistant • Hepatitis C (non-perinatal) • Shigellosis -

Purification and Characterization of a Shigella Dysenteriae 1- Like Toxin Produced by Escherichia Coli
CORE Metadata, citation and similar papers at core.ac.uk Provided by UNL | Libraries University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Uniformed Services University of the Health Sciences U.S. Department of Defense 1983 Purification and Characterization of a Shigella dysenteriae 1- Like Toxin Produced by Escherichia coli Alison D. O'Brien Uniformed Services University of the Health Sciences, [email protected] Gerald D. LaVeck Uniformed Services University of the Health Sciences Follow this and additional works at: https://digitalcommons.unl.edu/usuhs Part of the Medicine and Health Sciences Commons O'Brien, Alison D. and LaVeck, Gerald D., "Purification and Characterization of a Shigella dysenteriae 1- Like Toxin Produced by Escherichia coli" (1983). Uniformed Services University of the Health Sciences. 99. https://digitalcommons.unl.edu/usuhs/99 This Article is brought to you for free and open access by the U.S. Department of Defense at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Uniformed Services University of the Health Sciences by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. INFECTION AND IMMUNITY, May 1983, p. 675-683 Vol. 40, No. 2 0019-9567/83/050675-09$02.00/0 Copyright C 1983, American Society for Microbiology Purification and Characterization of a Shigella dysenteriae 1- Like Toxin Produced by Escherichia coli ALISON D. O'BRIEN* AND GERALD D. LAVECKt Department of Microbiology, Uniformed Services University of the Health Sciences, Bethesda, Maryland 20814 Received 10 January 1983/Accepted 18 February 1983 A toxin from an enteropathogenic strain of Escherichia coli (E. coli H30) was purified to apparent homogeneity from cell lysates. -

Communicable Diseases Weekly Report
Communicable Diseases Weekly Report Week 08, 21 February to 27 February 2021 In summary, we report: • Chancroid – one new case in a returned traveller • Rodent-borne disease risks • Novel coronavirus 2019 (COVID-19) • Summary of notifiable conditions activity in NSW For further information see NSW Health infectious diseases page. This includes links to other NSW Health infectious disease surveillance reports and a diseases data page for a range of notifiable infectious diseases. Chancroid One case of chancroid was notified this reporting week in a traveller returning from overseas. Chancroid is an acute sexually transmitted bacterial infection that causes painful genital ulcers. The condition is now rarely seen in Australia and only one other case has been notified in NSW during the past decade. Although the incidence of chancroid is decreasing globally, it is still reported in some regions within Africa, Asia, the Caribbean and South Pacific. Chancroid genital ulcer disease is a known risk factor for the transmission of HIV. The bacterium that causes chancroid, Haemophilus ducreyi, is usually transmitted through anal, oral, or vaginal sex with an infected person. After infection, one or more ulcers (sores) develop on the genitals or around the anus. Non-genital skin infections have also been reported globally, through non-sexual skin-to-skin contact with an infected person. The ulcers are usually painful, but rarely can be asymptomatic. Swelling in the groin (due to enlarged painful lymph nodes that can liquify and develop into buboes) can also occur. Other symptoms may include pain during sexual intercourse or while urinating. An infected person can spread the infection from their genital region to other parts of their body. -

Annual Summary of Communicable Disease Reported to MDH, 2003
MINNESOTA DEPARTMENT OF HEALTH DISEASE CONTROL N EWSLETTER Volume 32, Number 4 (pages 33-52) July/August 2004 Annual Summary of Communicable Diseases Reported to the Minnesota Department of Health, 2003 Introduction Minnesota Government Data Practices do not appear in Table 2 because the Assessment is a core public health Act (Section 13.38). Provisions of the influenza surveillance system is based function. Surveillance for communi- Health Insurance Portability and on reported outbreaks rather than on cable diseases is one type of ongoing Accountability Act (HIPAA) allow for individual cases. assessment activity. Epidemiologic routine communicable disease report- surveillance is the systematic collec- ing without patient authorization. Incidence rates in this report were tion, analysis, and dissemination of calculated using disease-specific health data for the planning, implemen- Since April 1995, MDH has participated numerator data collected by MDH and a tation, and evaluation of public health as one of the Emerging Infections standardized set of denominator data programs. The Minnesota Department Program (EIP) sites funded by the derived from U.S. Census data. of Health (MDH) collects disease Centers for Disease Control and Disease incidence may be categorized surveillance information on certain Prevention (CDC) and, through this as occurring within the seven-county communicable diseases for the program, has implemented active Twin Cities metropolitan area (Twin purposes of determining disease hospital- and laboratory-based surveil- Cities metropolitan area) or outside of it impact, assessing trends in disease lance for several conditions, including (Greater Minnesota). occurrence, characterizing affected selected invasive bacterial diseases populations, prioritizing disease control and food-borne diseases. Anaplasmosis efforts, and evaluating disease preven- Human anaplasmosis (HA) is the new tion strategies. -

Longitudinal Characterization of the Gut Bacterial and Fungal Communities in Yaks
Journal of Fungi Article Longitudinal Characterization of the Gut Bacterial and Fungal Communities in Yaks Yaping Wang 1,2,3, Yuhang Fu 3, Yuanyuan He 3, Muhammad Fakhar-e-Alam Kulyar 3 , Mudassar Iqbal 3,4, Kun Li 1,2,* and Jiaguo Liu 1,2,* 1 Institute of Traditional Chinese Veterinary Medicine, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing 210095, China; [email protected] 2 MOE Joint International Research Laboratory of Animal Health and Food Safety, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing 210095, China 3 College of Veterinary Medicine, Huazhong Agricultural University, Wuhan 430070, China; [email protected] (Y.F.); [email protected] (Y.H.); [email protected] (M.F.-e.-A.K.); [email protected] (M.I.) 4 Faculty of Veterinary and Animal Sciences, The Islamia University of Bahawalpur, Bahawalpur 63100, Pakistan * Correspondence: [email protected] (K.L.); [email protected] (J.L.) Abstract: Development phases are important in maturing immune systems, intestinal functions, and metabolism for the construction, structure, and diversity of microbiome in the intestine during the entire life. Characterizing the gut microbiota colonization and succession based on age-dependent effects might be crucial if a microbiota-based therapeutic or disease prevention strategy is adopted. The purpose of this study was to reveal the dynamic distribution of intestinal bacterial and fungal communities across all development stages in yaks. Dynamic changes (a substantial difference) in the structure and composition ratio of the microbial community were observed in yaks that Citation: Wang, Y.; Fu, Y.; He, Y.; matched the natural aging process from juvenile to natural aging. -

Insights Into the Pathogenicity of Burkholderia Pseudomallei
REVIEWS Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei W. Joost Wiersinga*, Tom van der Poll*, Nicholas J. White‡§, Nicholas P. Day‡§ and Sharon J. Peacock‡§ Abstract | Burkholderia pseudomallei is a potential bioterror agent and the causative agent of melioidosis, a severe disease that is endemic in areas of Southeast Asia and Northern Australia. Infection is often associated with bacterial dissemination to distant sites, and there are many possible disease manifestations, with melioidosis septic shock being the most severe. Eradication of the organism following infection is difficult, with a slow fever-clearance time, the need for prolonged antibiotic therapy and a high rate of relapse if therapy is not completed. Mortality from melioidosis septic shock remains high despite appropriate antimicrobial therapy. Prevention of disease and a reduction in mortality and the rate of relapse are priority areas for future research efforts. Studying how the disease is acquired and the host–pathogen interactions involved will underpin these efforts; this review presents an overview of current knowledge in these areas, highlighting key topics for evaluation. Melioidosis is a serious disease caused by the aerobic, rifamycins, colistin and aminoglycosides), but is usually Gram-negative soil-dwelling bacillus Burkholderia pseu- susceptible to amoxicillin-clavulanate, chloramphenicol, domallei and is most common in Southeast Asia and doxycycline, trimethoprim-sulphamethoxazole, ureido- Northern Australia. Melioidosis is responsible for 20% of penicillins, ceftazidime and carbapenems2,4. Treatment all community-acquired septicaemias and 40% of sepsis- is required for 20 weeks and is divided into intravenous related mortality in northeast Thailand. Reported cases are and oral phases2,4. Initial intravenous therapy is given likely to represent ‘the tip of the iceberg’1,2, as confirmation for 10–14 days; ceftazidime or a carbapenem are the of disease depends on bacterial isolation, a technique that drugs of choice. -

Plesiomonas Shigelloides S000142446 Serratia Plymuthica
0.01 Plesiomonas shigelloides S000142446 Serratia plymuthica S000016955 Serratia ficaria S000010445 Serratia entomophila S000015406 Leads to highlighted edge in Supplemental Phylogeny 1 Xenorhabdus hominickii S000735562 0.904 Xenorhabdus poinarii S000413914 0.998 0.868 Xenorhabdus griffiniae S000735553 Proteus myxofaciens S000728630 0.862 Proteus vulgaris S000004373 Proteus mirabilis S000728627 0.990 0.822 Arsenophonus nasoniae S000404964 OTU from Corby-Harris (Unpublished) #seqs-1 GQ988429 OTU from Corby-Harris et al (2007) #seqs-1 DQ980880 0.838 Providencia stuartii S000001277 Providencia rettgeri S000544654 0.999 Providencia vermicola S000544657 0.980 Isolate from Juneja and Lazzaro (2009)-EU587107 Isolate from Juneja and Lazzaro (2009)-EU587113 0.605 0.928 Isolate from Juneja and Lazzaro (2009)-EU587095 0.588 Isolate from Juneja and Lazzaro (2009)-EU587101 Providencia rustigianii S000544651 0.575 Providencia alcalifaciens S000127327 0.962 OTU XDS 099-#libs(1/0)-#seqs(1/0) OTU XYM 085-#libs(13/4)-#seqs(401/23) 0.620 OTU XDY 021-#libs(1/1)-#seqs(3/1) Providencia heimbachae S000544652 Morganella psychrotolerans S000721631 0.994 Morganella morganii S000721642 OTU ICF 062-#libs(0/1)-#seqs(0/7) Morganella morganii S000129416 0.990 Biostraticola tofi S000901778 0.783 Sodalis glossinidius S000436949 Dickeya dadantii S000570097 0.869 0.907 0.526 Brenneria rubrifaciens S000022162 0.229 Brenneria salicis S000381181 0.806 OTU SEC 066-#libs(0/1)-#seqs(0/1) Brenneria quercina S000005099 0.995 Pragia fontium S000022757 Budvicia aquatica S000020702 -

Use of the Diagnostic Bacteriology Laboratory: a Practical Review for the Clinician
148 Postgrad Med J 2001;77:148–156 REVIEWS Postgrad Med J: first published as 10.1136/pmj.77.905.148 on 1 March 2001. Downloaded from Use of the diagnostic bacteriology laboratory: a practical review for the clinician W J Steinbach, A K Shetty Lucile Salter Packard Children’s Hospital at EVective utilisation and understanding of the Stanford, Stanford Box 1: Gram stain technique University School of clinical bacteriology laboratory can greatly aid Medicine, 725 Welch in the diagnosis of infectious diseases. Al- (1) Air dry specimen and fix with Road, Palo Alto, though described more than a century ago, the methanol or heat. California, USA 94304, Gram stain remains the most frequently used (2) Add crystal violet stain. USA rapid diagnostic test, and in conjunction with W J Steinbach various biochemical tests is the cornerstone of (3) Rinse with water to wash unbound A K Shetty the clinical laboratory. First described by Dan- dye, add mordant (for example, iodine: 12 potassium iodide). Correspondence to: ish pathologist Christian Gram in 1884 and Dr Steinbach later slightly modified, the Gram stain easily (4) After waiting 30–60 seconds, rinse with [email protected] divides bacteria into two groups, Gram positive water. Submitted 27 March 2000 and Gram negative, on the basis of their cell (5) Add decolorising solvent (ethanol or Accepted 5 June 2000 wall and cell membrane permeability to acetone) to remove unbound dye. Growth on artificial medium Obligate intracellular (6) Counterstain with safranin. Chlamydia Legionella Gram positive bacteria stain blue Coxiella Ehrlichia Rickettsia (retained crystal violet).