Sympatric Parallel Diversification of Major Oak Clades in the Americas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
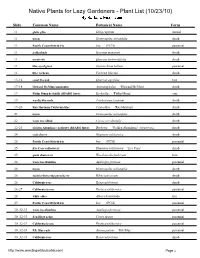
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

Seed Ecology Iii
SEED ECOLOGY III The Third International Society for Seed Science Meeting on Seeds and the Environment “Seeds and Change” Conference Proceedings June 20 to June 24, 2010 Salt Lake City, Utah, USA Editors: R. Pendleton, S. Meyer, B. Schultz Proceedings of the Seed Ecology III Conference Preface Extended abstracts included in this proceedings will be made available online. Enquiries and requests for hardcopies of this volume should be sent to: Dr. Rosemary Pendleton USFS Rocky Mountain Research Station Albuquerque Forestry Sciences Laboratory 333 Broadway SE Suite 115 Albuquerque, New Mexico, USA 87102-3497 The extended abstracts in this proceedings were edited for clarity. Seed Ecology III logo designed by Bitsy Schultz. i June 2010, Salt Lake City, Utah Proceedings of the Seed Ecology III Conference Table of Contents Germination Ecology of Dry Sandy Grassland Species along a pH-Gradient Simulated by Different Aluminium Concentrations.....................................................................................................................1 M Abedi, M Bartelheimer, Ralph Krall and Peter Poschlod Induction and Release of Secondary Dormancy under Field Conditions in Bromus tectorum.......................2 PS Allen, SE Meyer, and K Foote Seedling Production for Purposes of Biodiversity Restoration in the Brazilian Cerrado Region Can Be Greatly Enhanced by Seed Pretreatments Derived from Seed Technology......................................................4 S Anese, GCM Soares, ACB Matos, DAB Pinto, EAA da Silva, and HWM Hilhorst -

Native Trees of Georgia
1 NATIVE TREES OF GEORGIA By G. Norman Bishop Professor of Forestry George Foster Peabody School of Forestry University of Georgia Currently Named Daniel B. Warnell School of Forest Resources University of Georgia GEORGIA FORESTRY COMMISSION Eleventh Printing - 2001 Revised Edition 2 FOREWARD This manual has been prepared in an effort to give to those interested in the trees of Georgia a means by which they may gain a more intimate knowledge of the tree species. Of about 250 species native to the state, only 92 are described here. These were chosen for their commercial importance, distribution over the state or because of some unusual characteristic. Since the manual is intended primarily for the use of the layman, technical terms have been omitted wherever possible; however, the scientific names of the trees and the families to which they belong, have been included. It might be explained that the species are grouped by families, the name of each occurring at the top of the page over the name of the first member of that family. Also, there is included in the text, a subdivision entitled KEY CHARACTERISTICS, the purpose of which is to give the reader, all in one group, the most outstanding features whereby he may more easily recognize the tree. ACKNOWLEDGEMENTS The author wishes to express his appreciation to the Houghton Mifflin Company, publishers of Sargent’s Manual of the Trees of North America, for permission to use the cuts of all trees appearing in this manual; to B. R. Stogsdill for assistance in arranging the material; to W. -

Diversidad Del Género Quercus (Fagaceae) En México
Boletín de la Sociedad Botánica de México 75: 33-53, 2004 DOI: 10.17129/botsci.1692 Bol.Soc.Bot.Méx. 75: 33-53 (2004) SISTEMÁTICA Y FLORÍSTICA DIVERSIDAD DEL GÉNERO QUERCUS (FAGACEAE) EN MÉXICO SUSANA VALENCIA-A. Herbario de la Facultad de Ciencias (FCME), Departamento de Biología Comparada, Universidad Nacional Autónoma de México, Ciudad Universitaria, México, 04510, México D.F. Correo-e: [email protected] Resumen: Se presenta una lista preliminar de 161 especies del género Quercus para México, ubicadas en tres secciones: 76 en la sección Lobatae (encinos rojos), 81 en la sección Quercus (encinos blancos) y cuatro especies en la sección Protobalanus (encinos intermedios). Se calcula que 109 especies son endémicas del país, de las cuales 47 pertenecen a la sección Quercus, 61 a la sección Lobatae y una a Protobalanus. México comparte con Estados Unidos 33 especies del género, mientras que con Centroamérica com- parte 20. Los estados con mayor diversidad de especies son Oaxaca, Nuevo León, Jalisco, Chihuahua y Veracruz. Las especies con distribución más amplia en nuestro país son Q. candicans, Q. castanea, Q. crassifolia, Q. laeta, Q. microphylla, Q. obtusata y Q. rugosa. Altitudinalmente las especies de Quercus se desarrollan entre 0 y 3,500 m, pero son más frecuentes entre 1,000 y 3,000 m. El conocimiento del género Quercus en México es aún deficiente y se necesita realizar más estudios en torno a este importante género. P alabras clave: di versidad, encino, endemismo, México, Quercus. Abstract: This study presents a preliminary list of 161 species from the genus Quercus, all native to Mexico. -

Adenostoma Fasciculatum Profile to Postv2.Xlsx
I. SPECIES Adenostoma fasciculatum Hooker & Arnott NRCS CODE: ADFA Family: Rosaceae A. f. var. obtusifolium, Ron A. f. var. fasciculatum., Riverside Co., A. Montalvo, RCRCD Vanderhoff (Creative Order: Rosales Commons CC) Subclass: Rosidae Class: Magnoliopsida A. Subspecific taxa 1. Adenostoma fasciculatum var. fasciculatum Hook. & Arn. 1. ADFAF 2. A. f. var. obusifolium S. Watson 2. ADFAO 3. A. f. var. prostratum Dunkle 3. (no NRCS code) B. Synonyms 1. A. f. var. densifolium Eastw. 2. A. brevifolium Nutt. 3. none. Formerly included as part of A. f. var. f. C. Common name 1. chamise, common chamise, California greasewood, greasewood, chamiso (Painter 2016) 2. San Diego chamise (Calflora 2016) 3. prostrate chamise (Calflora 2016) Phylogenetic studies using molecular sequence data placedAdenostoma closest to Chamaebatiaria and D. Taxonomic relationships Sorbaria (Morgan et al. 1994, Potter et al. 2007) and suggest tentative placement in subfamily Spiraeoideae, tribe Sorbarieae (Potter et al. 2007). E. Related taxa in region Adenostoma sparsifolium Torrey, known as ribbon-wood or red-shanks is the only other species of Adenostoma in California. It is a much taller, erect to spreading shrub of chaparral vegetation, often 2–6 m tall and has a more restricted distribution than A. fasciculatum. It occurs from San Luis Obispo Co. south into Baja California. Red-shanks produces longer, linear leaves on slender long shoots rather than having leaves clustered on short shoots (lacks "fascicled" leaves). Its bark is cinnamon-colored and in papery layers that sheds in long ribbons. F. Taxonomic issues The Jepson eFlora and the FNA recognize A. f. var. prostratum but the taxon is not recognized by USDA PLANTS (2016). -

Quercus ×Coutinhoi Samp. Discovered in Australia Charlie Buttigieg
XXX International Oaks The Journal of the International Oak Society …the hybrid oak that time forgot, oak-rod baskets, pros and cons of grafting… Issue No. 25/ 2014 / ISSN 1941-2061 1 International Oaks The Journal of the International Oak Society … the hybrid oak that time forgot, oak-rod baskets, pros and cons of grafting… Issue No. 25/ 2014 / ISSN 1941-2061 International Oak Society Officers and Board of Directors 2012-2015 Officers President Béatrice Chassé (France) Vice-President Charles Snyers d’Attenhoven (Belgium) Secretary Gert Fortgens (The Netherlands) Treasurer James E. Hitz (USA) Board of Directors Editorial Committee Membership Director Chairman Emily Griswold (USA) Béatrice Chassé Tour Director Members Shaun Haddock (France) Roderick Cameron International Oaks Allen Coombes Editor Béatrice Chassé Shaun Haddock Co-Editor Allen Coombes (Mexico) Eike Jablonski (Luxemburg) Oak News & Notes Ryan Russell Editor Ryan Russell (USA) Charles Snyers d’Attenhoven International Editor Roderick Cameron (Uruguay) Website Administrator Charles Snyers d’Attenhoven For contributions to International Oaks contact Béatrice Chassé [email protected] or [email protected] 0033553621353 Les Pouyouleix 24800 St.-Jory-de-Chalais France Author’s guidelines for submissions can be found at http://www.internationaloaksociety.org/content/author-guidelines-journal-ios © 2014 International Oak Society Text, figures, and photographs © of individual authors and photographers. Graphic design: Marie-Paule Thuaud / www.lecentrecreatifducoin.com Photos. Cover: Charles Snyers d’Attenhoven (Quercus macrocalyx Hickel & A. Camus); p. 6: Charles Snyers d’Attenhoven (Q. oxyodon Miq.); p. 7: Béatrice Chassé (Q. acerifolia (E.J. Palmer) Stoynoff & W. J. Hess); p. 9: Eike Jablonski (Q. ithaburensis subsp. -

Key to Leaves of Eastern Native Oaks
FHTET-2003-01 January 2003 Front Cover: Clockwise from top left: white oak (Q. alba) acorns; willow oak (Q. phellos) leaves and acorns; Georgia oak (Q. georgiana) leaf; chinkapin oak (Q. muehlenbergii) acorns; scarlet oak (Q. coccinea) leaf; Texas live oak (Q. fusiformis) acorns; runner oak (Q. pumila) leaves and acorns; background bur oak (Q. macrocarpa) bark. (Design, D. Binion) Back Cover: Swamp chestnut oak (Q. michauxii) leaves and acorns. (Design, D. Binion) FOREST HEALTH TECHNOLOGY ENTERPRISE TEAM TECHNOLOGY TRANSFER Oak Identification Field Guide to Native Oak Species of Eastern North America John Stein and Denise Binion Forest Health Technology Enterprise Team USDA Forest Service 180 Canfield St., Morgantown, WV 26505 Robert Acciavatti Forest Health Protection Northeastern Area State and Private Forestry USDA Forest Service 180 Canfield St., Morgantown, WV 26505 United States Forest FHTET-2003-01 Department of Service January 2003 Agriculture NORTH AMERICA 100th Meridian ii iii ACKNOWLEDGMENTS The authors wish to thank all those who helped with this publication. We are grateful for permission to use the drawings illustrated by John K. Myers, Flagstaff, AZ, published in the Flora of North America, North of Mexico, vol. 3 (Jensen 1997). We thank Drs. Cynthia Huebner and Jim Colbert, U.S. Forest Service, Northeastern Research Station, Disturbance Ecology and Management of Oak-Dominated Forests, Morgantown, WV; Dr. Martin MacKenzie, U.S. Forest Service, Northeastern Area State and Private Forestry, Forest Health Protection, Morgantown, WV; Dr. Steven L. Stephenson, Department of Biology, Fairmont State College, Fairmont, WV; Dr. Donna Ford-Werntz, Eberly College of Arts and Sciences, West Virginia University, Morgantown, WV; Dr. -

Geology As a Georegional Influence on Quercus Fagaceae Distribution
GEOLOGY AS A GEOREGIONAL INFLUENCE ON Quercus FAGACEAE DISTRIBUTION IN DENTON AND COKE COUNTIES OF CENTRAL AND NORTH CENTRAL TEXAS AND CHOCTAW COUNTY OF SOUTHEASTERN OKLAHOMA, USING GIS AS AN ANALYTICAL TOOL George F. Maxey, B.S., M.S. Dissertation Prepared for the Degree of DOCTOR OF PHILOSOPHY UNIVERSITY OF NORTH TEXAS December 2007 APPROVED: C. Reid Ferring, Major Professor Miguel Avevedo, Committee Member Kenneth Dickson, Committee Member Donald Lyons, Committee Member Paul Hudak, Committee Member and Chair of the Department of Geography Sandra L. Terrell, Dean of the Robert B. Toulouse School of Graduate Studies Maxey, George F. Geology as a Georegional Influence on Quercus Fagaceae Distribution in Denton and Coke Counties of Central and North Central Texas and Choctaw County of Southeastern Oklahoma, Using GIS as an Analytical Tool. Doctor of Philosophy (Environmental Science), December 2007, 198 pp., 30 figures, 24 tables, references, 57 titles. This study elucidates the underlying relationships for the distribution of oak landcover on bedrock and soil orders in two counties in Texas and one in Oklahoma. ESRI’s ArcGis and ArcMap was used to create surface maps for Denton and Coke Counties, Texas and Choctaw County, Oklahoma. Attribute tables generated in GIS were exported into a spreadsheet software program and frequency tables were created for every formation and soil order in the tri-county research area. The results were both a visual and numeric distribution of oaks in the transition area between the eastern hardwood forests and the Great Plains. Oak distributions are changing on this transition area of the South Central Plains. -

Redalyc.Los Usos No Leñosos De Los Encinos En México
Boletín de la Sociedad Botánica de México ISSN: 0366-2128 [email protected] Sociedad Botánica de México México Luna José, Azucena de Lourdes; Montalvo Espinosa, Linda; Rendón Aguilar, Beatriz Los usos no leñosos de los encinos en México Boletín de la Sociedad Botánica de México, núm. 72, junio, 2003, pp. 107-117 Sociedad Botánica de México Distrito Federal, México Disponible en: http://www.redalyc.org/articulo.oa?id=57707204 Cómo citar el artículo Número completo Sistema de Información Científica Más información del artículo Red de Revistas Científicas de América Latina, el Caribe, España y Portugal Página de la revista en redalyc.org Proyecto académico sin fines de lucro, desarrollado bajo la iniciativa de acceso abierto Bol. Soc. Bot. Méx. 72: 107-117 (2003) BOTÁNICA ECONÓMICA Y ETNOBOTÁNICA LOS USOS NO LEÑOSOS DE LOS ENCINOS EN MÉXICO AZUCENA DE LOURDES LUNA-JOSÉ1, LINDA MONTALVO-ESPINOSA1 Y BEATRIZ RENDÓN-AGUILAR2 1Colegio de Posgraduados, Montecillos, México 2Departamento de Biología, Universidad Autónoma Metropolitana-Iztapalapa. A.P. 55-535. México, D.F. C.P. 09340. Fax 58-04-46-88. Correo-e: [email protected] Resumen: Se presenta una revisión bibliográfica y de herbario de los usos no leñosos de algunos encinos en México. Cincuenta y cinco especies de Quercus se utilizan y obtienen exclusivamente mediante la recolección. La mayoría se emplea en los estados ubicados en el centro y sur del país. No se encontró una relación entre el número de especies utilizadas y la diversidad de especies por estado. Se registraron cinco categorías de uso: (1) medicinal, que se relaciona principalmente con problemas del aparato digestivo; (2) alimenticio, que comprende la elaboración y consumo de alimento fresco o procesado; (3) artesanal, para la elaboración de diferentes artículos como rosarios o juguetes; (4) forraje, principalmente para la alimentación de ganado porcino y caprino; (5) taninos y colorantes, para curtir piel, como mordiente y para teñir hilo. -

Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- BIBLIOGRAPHY
Flora of the Carolinas, Virginia, and Georgia, Working Draft of 17 March 2004 -- BIBLIOGRAPHY BIBLIOGRAPHY Ackerfield, J., and J. Wen. 2002. A morphometric analysis of Hedera L. (the ivy genus, Araliaceae) and its taxonomic implications. Adansonia 24: 197-212. Adams, P. 1961. Observations on the Sagittaria subulata complex. Rhodora 63: 247-265. Adams, R.M. II, and W.J. Dress. 1982. Nodding Lilium species of eastern North America (Liliaceae). Baileya 21: 165-188. Adams, R.P. 1986. Geographic variation in Juniperus silicicola and J. virginiana of the Southeastern United States: multivariant analyses of morphology and terpenoids. Taxon 35: 31-75. ------. 1995. Revisionary study of Caribbean species of Juniperus (Cupressaceae). Phytologia 78: 134-150. ------, and T. Demeke. 1993. Systematic relationships in Juniperus based on random amplified polymorphic DNAs (RAPDs). Taxon 42: 553-571. Adams, W.P. 1957. A revision of the genus Ascyrum (Hypericaceae). Rhodora 59: 73-95. ------. 1962. Studies in the Guttiferae. I. A synopsis of Hypericum section Myriandra. Contr. Gray Herbarium Harv. 182: 1-51. ------, and N.K.B. Robson. 1961. A re-evaluation of the generic status of Ascyrum and Crookea (Guttiferae). Rhodora 63: 10-16. Adams, W.P. 1973. Clusiaceae of the southeastern United States. J. Elisha Mitchell Sci. Soc. 89: 62-71. Adler, L. 1999. Polygonum perfoliatum (mile-a-minute weed). Chinquapin 7: 4. Aedo, C., J.J. Aldasoro, and C. Navarro. 1998. Taxonomic revision of Geranium sections Batrachioidea and Divaricata (Geraniaceae). Ann. Missouri Bot. Gard. 85: 594-630. Affolter, J.M. 1985. A monograph of the genus Lilaeopsis (Umbelliferae). Systematic Bot. Monographs 6. Ahles, H.E., and A.E. -

Redalyc.Tree and Tree-Like Species of Mexico: Apocynaceae, Cactaceae
Revista Mexicana de Biodiversidad ISSN: 1870-3453 [email protected] Universidad Nacional Autónoma de México México Ricker, Martin; Valencia-Avalos, Susana; Hernández, Héctor M.; Gómez-Hinostrosa, Carlos; Martínez-Salas, Esteban M.; Alvarado-Cárdenas, Leonardo O.; Wallnöfer, Bruno; Ramos, Clara H.; Mendoza, Pilar E. Tree and tree-like species of Mexico: Apocynaceae, Cactaceae, Ebenaceae, Fagaceae, and Sapotaceae Revista Mexicana de Biodiversidad, vol. 87, núm. 4, diciembre, 2016, pp. 1189-1202 Universidad Nacional Autónoma de México Distrito Federal, México Available in: http://www.redalyc.org/articulo.oa?id=42548632003 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Available online at www.sciencedirect.com Revista Mexicana de Biodiversidad Revista Mexicana de Biodiversidad 87 (2016) 1189–1202 www.ib.unam.mx/revista/ Taxonomy and systematics Tree and tree-like species of Mexico: Apocynaceae, Cactaceae, Ebenaceae, Fagaceae, and Sapotaceae Especies arbóreas y arborescentes de México: Apocynaceae, Cactaceae, Ebenaceae, Fagaceae y Sapotaceae a,∗ b a a Martin Ricker , Susana Valencia-Avalos , Héctor M. Hernández , Carlos Gómez-Hinostrosa , a b c Esteban M. Martínez-Salas , Leonardo O. Alvarado-Cárdenas , Bruno Wallnöfer , a a Clara H. Ramos , Pilar E. Mendoza a Herbario Nacional de México (MEXU), Departamento -

Palinotaxonomía De Scrophulariaceae Sensu Lato
Bol. Soc. Argent. Bot. 51 (2) 2016 M. M. Sosa y C. R. Salgado - Palinotaxonomía de ScrophulariaceaeISSN sensu 0373-580 lato X Bol. Soc. Argent. Bot. 51 (2): 299-321. 2016 VALOR TAXONÓMICO DEL POLEN EN SCROPHULARIACEAE SENSU LATO MARÍA DE LAS MERCEDES SOSA1,3 y CRISTINA R. SALGADO2,3 Resumen: Este trabajo es el resultado del estudio palinomorfológico de 32 especies pertenecientes a 19 géneros de Scrophulariaceae sensu lato. Además se realizó un registro exhaustivo de la información bibliográfica existente sobre la morfología polínica con el objetivo de analizar las relaciones entre las variables palinológicas y la clasificación sistemática actual. Los granos de polen analizados son: mónades, radiosimétricos e isopolares, de tamaño variable desde pequeños (11 µm) hasta medianos (51 µm), predominantemente esferoidales (P/E= 1), a veces suboblatos (P/E= 0,76), oblato-esferoidales (P/E= 0,95), prolatos (P/E=1,63) y perprolatos (P/E= 2,63). Inaperturados, 3-(4-5) colpados, 3-(4-5) colporados, espiraperturados, 2-8 sincolpados, 3-diploporados y pantoporados. La exina puede ser semitectada (retipilada, reticulada o estriada perforada) y tectada (psilada, foveolada, perforada, escábrida, rugulada, verrugosa, granulada, estriada o equinulada). Se realizó un análisis estadístico en base a una matriz de datos, compuesta por los caracteres palinológicos de 32 especies estudiadas sumado a la recopilación bibliográfica de 140 géneros y 218 especies pertenecientes a Scrophulariaceae sensu lato. El análisis de agrupamiento (UPGMA), confirmó que las diferencias palinomorfológicas apoyan la clasificación sistemática actual. Palabras clave: Morfología del polen, sistemática, euripolínico, Scrophulariaceae. Summary: Taxonomic value of pollen in Scrophulariaceae sensu lato. This paper is result of a study of pollen morphology of 32 species belonging to 19 genera of Scrophulariaceae sensu lato.