From the Frying Pan: an Unusual Dwarf Shrub from Namibia Turns out to Be a New Brassicalean Family
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -
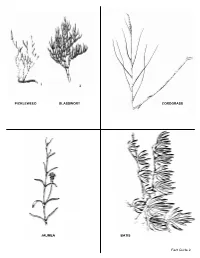
Plant Field Guide
2 PICKLEWEED GLASSWORT CORDGRASS JAUMEA BATIS Field Guide 9 PICKLEWEED Amaranth Family 3 kinds, 2 examples CORDGRASS Grass Family 1 Pickleweed Sarcocornia pacifica Spartina foliosa Glasswort Arthrocnemum subterminalis 2 HABITAT: Growns in the low marsh where the HABITAT: Found throughout the salt marsh. roots are continually bathed in ocean water. APPEARANCE: Stems look like a chain of small APPEARANCE: Look for a tall grass which is pickles. higher than the other plants in the salt marsh. REPRODUCTION: The flowers of all pickleweeds REPRODUCTION: All grasses are wind pollinated. are pollinated by the wind. The small flowers are Look for straw colored spikes of densely packed hard to see because they have no colorful petals flowers. Male flowers will have pollen and the female flowers will show graceful waving stigmas to ADAPTATION TO SALT: Pickleweeds are some of catch the pollen. the many marsh plants that use salt storage (they are accumulators). Also called succulents, these ADAPTATION TO SALT: All the salt marsh plants are swollen with the stored salty water. grasses are salt excreters using special pores to When the salt concentration becomes too high the push out droplets of salty water. Look on the grass cells will die. blades for salt crystals. See sea lavender. ECOLOGICAL RELATIONSHIPS: Frequently the ECOLOGICAL RELATIONSHIPS: Home for the most common plants in the marsh, they provide endangered bird, the Light-footed Clapper Rail. shelter and food for invertebrates. Belding’s A spider lives its whole life inside the blades. Savannah Sparrows build their nests in the Important food for grazing animals. glasswort. BATIS or SALTWORT Saltwort Family Batis maritima HABITAT: Most frequently found in the low marsh. -

A Compilation and Analysis of Food Plants Utilization of Sri Lankan Butterfly Larvae (Papilionoidea)
MAJOR ARTICLE TAPROBANICA, ISSN 1800–427X. August, 2014. Vol. 06, No. 02: pp. 110–131, pls. 12, 13. © Research Center for Climate Change, University of Indonesia, Depok, Indonesia & Taprobanica Private Limited, Homagama, Sri Lanka http://www.sljol.info/index.php/tapro A COMPILATION AND ANALYSIS OF FOOD PLANTS UTILIZATION OF SRI LANKAN BUTTERFLY LARVAE (PAPILIONOIDEA) Section Editors: Jeffrey Miller & James L. Reveal Submitted: 08 Dec. 2013, Accepted: 15 Mar. 2014 H. D. Jayasinghe1,2, S. S. Rajapaksha1, C. de Alwis1 1Butterfly Conservation Society of Sri Lanka, 762/A, Yatihena, Malwana, Sri Lanka 2 E-mail: [email protected] Abstract Larval food plants (LFPs) of Sri Lankan butterflies are poorly documented in the historical literature and there is a great need to identify LFPs in conservation perspectives. Therefore, the current study was designed and carried out during the past decade. A list of LFPs for 207 butterfly species (Super family Papilionoidea) of Sri Lanka is presented based on local studies and includes 785 plant-butterfly combinations and 480 plant species. Many of these combinations are reported for the first time in Sri Lanka. The impact of introducing new plants on the dynamics of abundance and distribution of butterflies, the possibility of butterflies being pests on crops, and observations of LFPs of rare butterfly species, are discussed. This information is crucial for the conservation management of the butterfly fauna in Sri Lanka. Key words: conservation, crops, larval food plants (LFPs), pests, plant-butterfly combination. Introduction Butterflies go through complete metamorphosis 1949). As all herbivorous insects show some and have two stages of food consumtion. -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Arthur Monrad Johnson Colletion of Botanical Drawings
http://oac.cdlib.org/findaid/ark:/13030/kt7489r5rb No online items Arthur Monrad Johnson colletion of botanical drawings 1914-1941 Processed by Pat L. Walter. Louise M. Darling Biomedical Library History and Special Collections Division History and Special Collections Division UCLA 12-077 Center for Health Sciences Box 951798 Los Angeles, CA 90095-1798 Phone: 310/825-6940 Fax: 310/825-0465 Email: [email protected] URL: http://www.library.ucla.edu/libraries/biomed/his/ ©2008 The Regents of the University of California. All rights reserved. Arthur Monrad Johnson colletion 48 1 of botanical drawings 1914-1941 Descriptive Summary Title: Arthur Monrad Johnson colletion of botanical drawings, Date (inclusive): 1914-1941 Collection number: 48 Creator: Johnson, Arthur Monrad 1878-1943 Extent: 3 boxes (2.5 linear feet) Repository: University of California, Los Angeles. Library. Louise M. Darling Biomedical Library History and Special Collections Division Los Angeles, California 90095-1490 Abstract: Approximately 1000 botanical drawings, most in pen and black ink on paper, of the structural parts of angiosperms and some gymnosperms, by Arthur Monrad Johnson. Many of the illustrations have been published in the author's scientific publications, such as his "Taxonomy of the Flowering Plants" and articles on the genus Saxifraga. Dr. Johnson was both a respected botanist and an accomplished artist beyond his botanical subjects. Physical location: Collection stored off-site (Southern Regional Library Facility): Advance notice required for access. Language of Material: Collection materials in English Preferred Citation [Identification of item], Arthur Monrad Johnson colletion of botanical drawings (Manuscript collection 48). Louise M. Darling Biomedical Library History and Special Collections Division, University of California, Los Angeles. -

Medicinal Practices of Sacred Natural Sites: a Socio-Religious Approach for Successful Implementation of Primary
Medicinal practices of sacred natural sites: a socio-religious approach for successful implementation of primary healthcare services Rajasri Ray and Avik Ray Review Correspondence Abstract Rajasri Ray*, Avik Ray Centre for studies in Ethnobiology, Biodiversity and Background: Sacred groves are model systems that Sustainability (CEiBa), Malda - 732103, West have the potential to contribute to rural healthcare Bengal, India owing to their medicinal floral diversity and strong social acceptance. *Corresponding Author: Rajasri Ray; [email protected] Methods: We examined this idea employing ethnomedicinal plants and their application Ethnobotany Research & Applications documented from sacred groves across India. A total 20:34 (2020) of 65 published documents were shortlisted for the Key words: AYUSH; Ethnomedicine; Medicinal plant; preparation of database and statistical analysis. Sacred grove; Spatial fidelity; Tropical diseases Standard ethnobotanical indices and mapping were used to capture the current trend. Background Results: A total of 1247 species from 152 families Human-nature interaction has been long entwined in has been documented for use against eighteen the history of humanity. Apart from deriving natural categories of diseases common in tropical and sub- resources, humans have a deep rooted tradition of tropical landscapes. Though the reported species venerating nature which is extensively observed are clustered around a few widely distributed across continents (Verschuuren 2010). The tradition families, 71% of them are uniquely represented from has attracted attention of researchers and policy- any single biogeographic region. The use of multiple makers for its impact on local ecological and socio- species in treating an ailment, high use value of the economic dynamics. Ethnomedicine that emanated popular plants, and cross-community similarity in from this tradition, deals health issues with nature- disease treatment reflects rich community wisdom to derived resources. -

Ethnobotanical Study of Wild Edible Plants of Kara And
Teklehaymanot and Giday Journal of Ethnobiology and Ethnomedicine 2010, 6:23 http://www.ethnobiomed.com/content/6/1/23 JOURNAL OF ETHNOBIOLOGY AND ETHNOMEDICINE RESEARCH Open Access Ethnobotanical study of wild edible plants of Kara and Kwego semi-pastoralist people in Lower Omo River Valley, Debub Omo Zone, SNNPR, Ethiopia Tilahun Teklehaymanot*, Mirutse Giday Abstract Background: The rural populations in Ethiopia have a rich knowledge of wild edible plants and consumption of wild edible plants is still an integral part of the different cultures in the country. In the southern part of the country, wild edible plants are used as dietary supplements and a means of survival during times of food shortage. Therefore, the aim of this study is to document the wild edible plants gathered and consumed by Kara and Kwego people, and to analyze patterns of use between the two people. Methods: A cross sectional ethnobotanical study of wild edible plant species was conducted from January 2005 to March 2007. About 10% of each people: 150 Kara and 56 Kwego were randomly selected to serve as informants. Data were collected using semi-structured questionnaire and group discussions. Analysis of variance (a = 0.05) was used to test the similarity of species richness of wild edible plants reported by Kara and Kwego people; Pearson’sChi-squaretest(a =0.05) was used to test similarity of growth forms and plant parts of wild edible plants used between the two people. Results: Thirty-eight wild plant species were reported as food sources that were gathered and consumed both at times of plenty and scarcity; three were unique to Kara, five to Kwego and 14 had similar local names. -

In Chemistry, Glycosides Are Certain Molecules in Which a Sugar Part Is
GLYCOSIDES Glycosides may be defined as the organic compounds from plants or animal sources, which on enzymatic or acid hydrolysis give one or more sugar moieties along with non- sugar moiety. Glycosides play numerous important roles in living organisms. Many plants store important chemicals in the form of inactive glycosides; if these chemicals are needed, the glycosides are brought in contact with water and an enzyme, and the sugar part is broken off, making the chemical available for use. Many such plant glycosides are used as medications. In animals (including humans), poisons are often bound to sugar molecules in order to remove them from the body. Formally, a glycoside is any molecule in which a sugar group is bonded through its carbon atom to another group via an O-glycosidic bond or an S-glycosidic bond; glycosides involving the latter are also called thioglycosides. The sugar group is then known as the glycone and the non-sugar group as the aglycone or genin part of the glycoside. The glycone can consist of a single sugar group (monosaccharide) or several sugar groups (oligosaccharide). Classification Classification based on linkages Based on the linkage of sugar moiety to aglycone part 1. O-Glycoside:-Here the sugar is combined with alcoholic or phenolic hydroxyl function of aglycone.eg:-digitalis. 2. N-glycosides:-Here nitrogen of amino group is condensed with a sugar ,eg- Nucleoside 3. S-glycoside:-Here sugar is combined with sulphur of aglycone,eg- isothiocyanate glycosides. 4. C-glycosides:-By condensation of a sugar with a cabon atom, eg-Cascaroside, aloin. Glycosides can be classified by the glycone, by the type of glycosidic bond, and by the aglycone. -

Evolutionary History of Floral Key Innovations in Angiosperms Elisabeth Reyes
Evolutionary history of floral key innovations in angiosperms Elisabeth Reyes To cite this version: Elisabeth Reyes. Evolutionary history of floral key innovations in angiosperms. Botanics. Université Paris Saclay (COmUE), 2016. English. NNT : 2016SACLS489. tel-01443353 HAL Id: tel-01443353 https://tel.archives-ouvertes.fr/tel-01443353 Submitted on 23 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLS489 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY, préparée à l’Université Paris-Sud ÉCOLE DOCTORALE N° 567 Sciences du Végétal : du Gène à l’Ecosystème Spécialité de Doctorat : Biologie Par Mme Elisabeth Reyes Evolutionary history of floral key innovations in angiosperms Thèse présentée et soutenue à Orsay, le 13 décembre 2016 : Composition du Jury : M. Ronse de Craene, Louis Directeur de recherche aux Jardins Rapporteur Botaniques Royaux d’Édimbourg M. Forest, Félix Directeur de recherche aux Jardins Rapporteur Botaniques Royaux de Kew Mme. Damerval, Catherine Directrice de recherche au Moulon Président du jury M. Lowry, Porter Curateur en chef aux Jardins Examinateur Botaniques du Missouri M. Haevermans, Thomas Maître de conférences au MNHN Examinateur Mme. Nadot, Sophie Professeur à l’Université Paris-Sud Directeur de thèse M. -

Toxicity of Glucosinolates and Their Enzymatic Decomposition Products to Caenorhabditis Elegans
Journal of Nematology 27(3):258-262. 1995. © The Society of Nematologists 1995. Toxicity of Glucosinolates and Their Enzymatic Decomposition Products to Caenorhabditis elegans STEVEN G. DONKIN, 1 MARK A. EITEMAN, 2 AND PHILLIP L. WILLIAMS 1'3 Abstract: An aquatic 24-hour lethality test using Caenorhabditis elegans was used to assess toxicity of glucosinolates and their enzymatic breakdown products. In the absence of the enzyme thioglucosi- dase (myrosinase), allyl glucosinolate (sinigrin) was found to be nontoxic at all concentrations tested, while a freeze-dried, dialyzed water extract of Crambe abyssinica containing 26% 2-hydroxyl 3-butenyl glucosinolate (epi-progoitrin) had a 50% lethal concentration (LC50) of 18.5 g/liter. Addition of the enzyme increased the toxicity (LCs0 value) of sinigrin to 0.5 g/liter, but the enzyme had no effect on the toxicity of the C. abyssinica extract. Allyl isothiocyanate and allyl cyanide, two possible breakdown products of sinigrin, had an LC50 value of 0.04 g/liter and approximately 3 g/liter, respectively. Liquid chromatographic studies showed that a portion of the sinigrin decomposed into allyl isothio- cyanate. The resuhs indicated that allyl isothiocyanate is nearly three orders of magnitude more toxic to C. elegans than the corresponding glncosinolate, suggesting isothiocyanate formation would im- prove nematode control from application of glucosinolates. Key words: Caenorhabditis elegans, Crambe abyssinica, enzyme, epi-progoitrin, glucosinolate, myrosi- nase, physiology, sinigrin, thioglucosidase. Glucosinolates are naturally occurring position products or between decomposi- compounds found primarily in plants of tion products. the family Cruciferae, where they are An objective of this work was to quantify thought to serve as repellents to potential the toxicity to the free-living nematode pests (5,10). -

Full of Beans: a Study on the Alignment of Two Flowering Plants Classification Systems
Full of beans: a study on the alignment of two flowering plants classification systems Yi-Yun Cheng and Bertram Ludäscher School of Information Sciences, University of Illinois at Urbana-Champaign, USA {yiyunyc2,ludaesch}@illinois.edu Abstract. Advancements in technologies such as DNA analysis have given rise to new ways in organizing organisms in biodiversity classification systems. In this paper, we examine the feasibility of aligning two classification systems for flowering plants using a logic-based, Region Connection Calculus (RCC-5) ap- proach. The older “Cronquist system” (1981) classifies plants using their mor- phological features, while the more recent Angiosperm Phylogeny Group IV (APG IV) (2016) system classifies based on many new methods including ge- nome-level analysis. In our approach, we align pairwise concepts X and Y from two taxonomies using five basic set relations: congruence (X=Y), inclusion (X>Y), inverse inclusion (X<Y), overlap (X><Y), and disjointness (X!Y). With some of the RCC-5 relationships among the Fabaceae family (beans family) and the Sapindaceae family (maple family) uncertain, we anticipate that the merging of the two classification systems will lead to numerous merged solutions, so- called possible worlds. Our research demonstrates how logic-based alignment with ambiguities can lead to multiple merged solutions, which would not have been feasible when aligning taxonomies, classifications, or other knowledge or- ganization systems (KOS) manually. We believe that this work can introduce a novel approach for aligning KOS, where merged possible worlds can serve as a minimum viable product for engaging domain experts in the loop. Keywords: taxonomy alignment, KOS alignment, interoperability 1 Introduction With the advent of large-scale technologies and datasets, it has become increasingly difficult to organize information using a stable unitary classification scheme over time. -

North Gujarat
Life Sciences Leaflets FREE DOWNLOAD ISSN 2277-4297(Print)0976–1098(Online) EXTRACTION OF SALVADORA OLEOIDES AND ITS PERFORMANCE ALONG WITH ANTIBIOTICS FOR ANTIMICROBIAL ACTIVITIES BHARGAV DAVE1, PIYUSH VYAS1*, MADHU PATEL2 AND 3 Universal Impact NAINESH PATEL Factor 0.9285:2012; 1. 1.2210:2013 SHETH M.N SCIENCE COLLEGE, PATAN. Index Copernicus 2. NAVSARI AGRICULTURAL UNIVERSITY, SURAT. ICV 2011: 5.09, 2012: 6.42, 2013: 3. PACIFIC UNIVERSITY, UDAIPUR, RAJASTHAN. 15.8, 2014:89.16 , 2015:78.30 Corresponding author’s e-mail: [email protected] NAAS Rating 2012 : 1.3; 2013-16:2.69 ABSTRACT: 2017: 3.98 Nature has given various ways to maintain people’s health. One way is to SJIF 2012: 3.947, 2013: 4.802 use herbal medicine. Herbal medicines have been used to treat various types Infobase Index 2015:4.56 of diseases for long times. The people are more attracting towards the use of Cosmos Impact Factor herbal drugs to cure various types of diseases. For treatment of several 2015: 4.366 diseases of human beings, plant drug ‘rasayana’ has always played a vital Received on: role. According to World health organization (WHO) more than 80% of the 6th December 2017 Revised on: world population is dependent on traditional medicine for their primary th 10 December 2017 health care needs.1 Accepted on: 12th December 2017 Herbal medicine, also called botanical medicine or phytomedicine, refers to Published on: the use of a plant’s seeds, barriers, roots, leaves, bark, or flowers for 1st January 2018 medicinal purposes. Long practiced outside of conventional medicine, Volume No.