PSI Biology Classification Multiple Choice
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Revised Glossary for AQA GCSE Biology Student Book
Biology Glossary amino acids small molecules from which proteins are A built abiotic factor physical or non-living conditions amylase a digestive enzyme (carbohydrase) that that affect the distribution of a population in an breaks down starch ecosystem, such as light, temperature, soil pH anaerobic respiration respiration without using absorption the process by which soluble products oxygen of digestion move into the blood from the small intestine antibacterial chemicals chemicals produced by plants as a defence mechanism; the amount abstinence method of contraception whereby the produced will increase if the plant is under attack couple refrains from intercourse, particularly when an egg might be in the oviduct antibiotic e.g. penicillin; medicines that work inside the body to kill bacterial pathogens accommodation ability of the eyes to change focus antibody protein normally present in the body acid rain rain water which is made more acidic by or produced in response to an antigen, which it pollutant gases neutralises, thus producing an immune response active site the place on an enzyme where the antimicrobial resistance (AMR) an increasing substrate molecule binds problem in the twenty-first century whereby active transport in active transport, cells use energy bacteria have evolved to develop resistance against to transport substances through cell membranes antibiotics due to their overuse against a concentration gradient antiretroviral drugs drugs used to treat HIV adaptation features that organisms have to help infections; they -
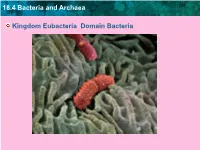
18.4 Bacteria and Archaea Kingdom Eubacteria Domain Bacteria
18.4 Bacteria and Archaea Kingdom Eubacteria Domain Bacteria 18.4 Bacteria and Archaea Description Bacteria are single-celled prokaryotes. 18.4 Bacteria and Archaea Where do they live? Prokaryotes are widespread on Earth. ( Est. over 1 billion types of bacteria, and over 1030 individual prokaryote cells on earth.) Found in all land and ocean environments, even inside other organisms! 18.4 Bacteria and Archaea Common Examples • E. Coli • Tetanus bacteria • Salmonella bacteria • Tuberculosis bacteria • Staphylococcus • Streptococcus 18.4 Bacteria and Archaea Modes Of Nutrition • Bacteria may be heterotrophs or autotrophs 18.4 Bacteria and Archaea Bacteria Reproduce How? • by binary fission. • exchange genes during conjugation= conjugation bridge increases diversity. • May survive by forming endospores = specialized cell with thick protective cell wall. TEM; magnification 6000x • Can survive for centuries until environment improves. Have been found in mummies! 18.4 Bacteria and Archaea • Bacteria Diagram – plasmid = small piece of genetic material, can replicate independently of the chromosome – flagellum = different than in eukaryotes, but for movement – pili = used to stick the bacteria to eachpili other or surfaces plasma membrance flagellum chromosome cell wall plasmid This diagram shows the typical structure of a prokaryote. Archaea and bacteria look very similar, although they have important molecular differences. 18.4 Bacteria and Archaea • Classified by: their need for oxygen, how they gram stain, and their shapes 18.4 Bacteria and Archaea Main Groups by Shapes – rod-shaped, called bacilli – spiral, called spirilla or spirochetes – spherical, called cocci Spirochaeta: spiral Lactobacilli: rod-shaped Enterococci: spherical 18.4 Bacteria and Archaea • Main Groups by their need for oxygen. -
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

And According to Bailey & (1943)
BLUMEA 24 (1978) 521—525 The Winteraceae pf the Old World. III. Notes on the ovary of Takhtajania W. Vink Rijksherbarium, Leiden, Netherlands INTRODUCTION Very recently Baranova & Leroy (Leroy, 1978) published a new genus Takhta- jania to accomodate the aberrant Bubbia perrieri Capuron. The outstanding characters of this genus are the anomocytic stomatal apparatus (Baranova, 1972; Bongers, 1973) and the unilocular bicarpellate ovary (Leroy, 1977, 1978). work the Winteraceae, also studied the of Bub- During my on I single specimen bia perrieri in existence. The late Capuron told me that he had tried to collect additional specimens but that he had not succeeded in doing so; according to him the type locality was completely deforested. In view of the scarcity of the material it was considered relevant to publish some additional notes without delay. MATERIAL AND METHODS I studied three ovaries of the type specimen (Perrier de la Bdthie 10158). One was cut longitudinally, slightly outside the plane of symmetry; another was cut and its half These drawn transversally upper again longitudinally. parts were (figs. 2 and 3) and then cleared according to Bailey & Nast (1943) (figs 5 and third boiled and then sectioned 6). The ovary was serially (fig. 4). OBSERVATIONS One of the ovaries was taken from a flowerbud of which the closed petals were 57 mm high. The diagram of this flowerbud is drawn in fig. 1. The 2 slightly ruptured calyx showed three putative original apices ± alternating with the three bracteoles at the base of the pedicel. The petals are all free. The ovary is its and the flattened, has a longitudinal groove on narrow sides, highest sta- mens are inserted adjoining its broadest sides. -

Biology Chapter 19 Kingdom Protista Domain Eukarya Description Kingdom Protista Is the Most Diverse of All the Kingdoms
Biology Chapter 19 Kingdom Protista Domain Eukarya Description Kingdom Protista is the most diverse of all the kingdoms. Protists are eukaryotes that are not animals, plants, or fungi. Some unicellular, some multicellular. Some autotrophs, some heterotrophs. Some with cell walls, some without. Didinium protist devouring a Paramecium protist that is longer than it is! Read about it on p. 573! Where Do They Live? • Because of their diversity, we find protists in almost every habitat where there is water or at least moisture! Common Examples • Ameba • Algae • Paramecia • Water molds • Slime molds • Kelp (Sea weed) Classified By: (DON’T WRITE THIS DOWN YET!!! • Mode of nutrition • Cell walls present or not • Unicellular or multicellular Protists can be placed in 3 groups: animal-like, plantlike, or funguslike. Didinium, is a specialist, only feeding on Paramecia. They roll into a ball and form cysts when there is are no Paramecia to eat. Paramecia, on the other hand are generalists in their feeding habits. Mode of Nutrition Depends on type of protist (see Groups) Main Groups How they Help man How they Hurt man Ecosystem Roles KEY CONCEPT Animal-like protists = PROTOZOA, are single- celled heterotrophs that can move. Oxytricha Reproduce How? • Animal like • Unicellular – by asexual reproduction – Paramecium – does conjugation to exchange genetic material Animal-like protists Classified by how they move. macronucleus contractile vacuole food vacuole oral groove micronucleus cilia • Protozoa with flagella are zooflagellates. – flagella help zooflagellates swim – more than 2000 zooflagellates • Some protists move with pseudopods = “false feet”. – change shape as they move –Ex. amoebas • Some protists move with pseudopods. -

Reconstructing the Basal Angiosperm Phylogeny: Evaluating Information Content of Mitochondrial Genes
55 (4) • November 2006: 837–856 Qiu & al. • Basal angiosperm phylogeny Reconstructing the basal angiosperm phylogeny: evaluating information content of mitochondrial genes Yin-Long Qiu1, Libo Li, Tory A. Hendry, Ruiqi Li, David W. Taylor, Michael J. Issa, Alexander J. Ronen, Mona L. Vekaria & Adam M. White 1Department of Ecology & Evolutionary Biology, The University Herbarium, University of Michigan, Ann Arbor, Michigan 48109-1048, U.S.A. [email protected] (author for correspondence). Three mitochondrial (atp1, matR, nad5), four chloroplast (atpB, matK, rbcL, rpoC2), and one nuclear (18S) genes from 162 seed plants, representing all major lineages of gymnosperms and angiosperms, were analyzed together in a supermatrix or in various partitions using likelihood and parsimony methods. The results show that Amborella + Nymphaeales together constitute the first diverging lineage of angiosperms, and that the topology of Amborella alone being sister to all other angiosperms likely represents a local long branch attrac- tion artifact. The monophyly of magnoliids, as well as sister relationships between Magnoliales and Laurales, and between Canellales and Piperales, are all strongly supported. The sister relationship to eudicots of Ceratophyllum is not strongly supported by this study; instead a placement of the genus with Chloranthaceae receives moderate support in the mitochondrial gene analyses. Relationships among magnoliids, monocots, and eudicots remain unresolved. Direct comparisons of analytic results from several data partitions with or without RNA editing sites show that in multigene analyses, RNA editing has no effect on well supported rela- tionships, but minor effect on weakly supported ones. Finally, comparisons of results from separate analyses of mitochondrial and chloroplast genes demonstrate that mitochondrial genes, with overall slower rates of sub- stitution than chloroplast genes, are informative phylogenetic markers, and are particularly suitable for resolv- ing deep relationships. -

Early Vessel Evolution and the Diverisification
American Journal of Botany 97(1): 80–93. 2010. E ARLY VESSEL EVOLUTION AND THE DIVERISIFICATION OF WOOD FUNCTION: INSIGHTS FROM MALAGASY CANELLALES 1 Patrick J. Hudson 2 , Jacqueline Razanatsoa 3 , and Taylor S. Feild 2,4 2 Department of Ecology and Evolutionary Biology, University of Tennessee, Knoxville, Tennessee 37919 USA; and 3 Herbier du Parc Botanique et Zoologique de Tsimbazaza, Rue Fernand, Kasanga, Tsimbazaza, Antananarivo 101, Madagascar Xylem vessels have long been proposed as a key innovation for the ecological diversifi cation of angiosperms by providing a breakthrough in hydraulic effi ciency to support high rates of photosynthesis and growth. However, recent studies demonstrated that angiosperm woods with structurally “ primitive ” vessels did not have greater whole stem hydraulic capacities as compared to vesselless angiosperms. As an alternative to the hydraulic superiority hypothesis, the heteroxylly hypothesis proposes that subtle hydraulic effi ciencies of primitive vessels over tracheids enabled new directions of functional specialization in the wood. However, the functional properties of early heteroxyllous wood remain unknown. We selected the two species of Canellales from Madagascar to test the heteroxylly hypothesis because Canellaceae (represented by Cinnamosma madagascariensis ) produces wood with vessels of an ancestral form, while Winteraceae, the sister clade (represented by Takhtajania perrieri) is vesselless. We found that heteroxy- lly correlated with increased wood functional diversity related predominantly to biomechanical specialization. However, vessels were not associated with greater stem hydraulic effi ciency or increased shoot hydraulic capacity. Our results support the heteroxylly hypothesis and highlight the importance integrating a broader ecological context to understand the evolution of vessels. Key words: Canellales; heteroxylly; hydraulic conductivity; shade tolerance; xylem evolution; vessel development. -

Structural Biology of the C-Terminal Domain Of
STRUCTURAL BIOLOGY OF THE C-TERMINAL DOMAIN OF EUKARYOTIC REPLICATION FACTOR MCM10 By Patrick David Robertson Dissertation Submitted to the Faculty of the Graduate School of Vanderbilt University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biological Sciences August, 2010 Nashville, Tennessee Approved: Brandt F. Eichman Walter J. Chazin James G. Patton Hassane Mchaourab To my wife Sabrina, thank you for your enduring love and support ii ACKNOWLEDGMENTS I would like to begin by expressing my sincerest gratitude to my mentor and Ph.D. advisor, Dr. Brandt Eichman. In addition to your excellent guidance and training, your passion for science has been a source of encouragement and inspiration over the past five years. I consider working with you to be a great privilege and I am grateful for the opportunity. I would also like to thank the members of my thesis committee: Drs. Walter Chazin, Ellen Fanning, James Patton, and Hassane Mchaourab. My research and training would not have been possible without your insight, advice and intellectual contributions. I would like to acknowledge all of the members of the Eichman laboratory, past and present, for their technical assistance and camaraderie over the years. I would especially like to thank Dr. Eric Warren for his contributions to the research presented here, as well for his friendship of the years. I would also like to thank Drs. Benjamin Chagot and Sivaraja Vaithiyalingam from the Chazin laboratory for their expert assistance and NMR training. I would especially like to thank my parents, Joyce and David, my brother Jeff, and the rest of my family for their love and encouragement throughout my life. -

BIRDS AS MARINE ORGANISMS: a REVIEW Calcofi Rep., Vol
AINLEY BIRDS AS MARINE ORGANISMS: A REVIEW CalCOFI Rep., Vol. XXI, 1980 BIRDS AS MARINE ORGANISMS: A REVIEW DAVID G. AINLEY Point Reyes Bird Observatory Stinson Beach, CA 94970 ABSTRACT asociadas con esos peces. Se indica que el estudio de las Only 9 of 156 avian families are specialized as sea- aves marinas podria contribuir a comprender mejor la birds. These birds are involved in marine energy cycles dinamica de las poblaciones de peces anterior a la sobre- during all aspects of their lives except for the 10% of time explotacion por el hombre. they spend in some nesting activities. As marine organ- isms their occurrence and distribution are directly affected BIRDS AS MARINE ORGANISMS: A REVIEW by properties of their oceanic habitat, such as water temp- As pointed out by Sanger(1972) and Ainley and erature, salinity, and turbidity. In their trophic relation- Sanger (1979), otherwise comprehensive reviews of bio- ships, almost all are secondary or tertiary carnivores. As logical oceanography have said little or nothing about a group within specific ecosystems, estimates of their seabirds in spite of the fact that they are the most visible feeding rates range between 20 and 35% of annual prey part of the marine biota. The reasons for this oversight are production. Their usual prey are abundant, schooling or- no doubt complex, but there are perhaps two major ones. ganisms such as euphausiids and squid (invertebrates) First, because seabirds have not been commercially har- and clupeids, engraulids, and exoccetids (fish). Their high vested to any significant degree, fisheries research, which rates of feeding and metabolism, and the large amounts of supplies most of our knowledge about marine ecosys- nutrients they return to the marine environment, indicate tems, has ignored them. -

Anatomy and Go Fish! Background
Anatomy and Go Fish! Background Introduction It is important to properly identify fi sh for many reasons: to follow the rules and regulations, for protection against sharp teeth or protruding spines, for the safety of the fi sh, and for consumption or eating purposes. When identifying fi sh, scientists and anglers use specifi c vocabulary to describe external or outside body parts. These body parts are common to most fi sh. The difference in the body parts is what helps distinguish one fi sh from another, while their similarities are used to classify them into groups. There are approximately 29,000 fi sh species in the world. In order to identify each type of fi sh, scientists have grouped them according to their outside body parts, specifi cally the number and location of fi ns, and body shape. Classifi cation Using a system of classifi cation, scientists arrange all organisms into groups based on their similarities. The fi rst system of classifi cation was proposed in 1753 by Carolus Linnaeus. Linnaeus believed that each organism should have a binomial name, genus and species, with species being the smallest organization unit of life. Using Linnaeus’ system as a guide, scientists created a hierarchical system known as taxonomic classifi cation, in which organisms are classifi ed into groups based on their similarities. This hierarchical system moves from largest and most general to smallest and most specifi c: kingdom, phylum, class, order, family, genus, and species. {See Figure 1. Taxonomic Classifi cation Pyramid}. For example, fi sh belong to the kingdom Animalia, the phylum Chordata, and from there are grouped more specifi cally into several classes, orders, families, and thousands of genus and species. -

A Knowledge-Centric E-Research Platform for Marine Life and Oceanographic Research
A KNOWLEDGE-CENTRIC E-RESEARCH PLATFORM FOR MARINE LIFE AND OCEANOGRAPHIC RESEARCH Ali Daniyal, Samina Abidi, Ashraf AbuSharekh, Mei Kuan Wong and S. S. R. Abidi Department of Computer Science, Dalhousie University, 6050 Univeristy Ave, Halifax, Canada Keywords: e-Research, Knowledge management, Web services, Marine life, Oceanography. Abstract: In this paper we present a knowledge centric e-Research platform to support collaboration between two diverse scientific communities—i.e. Oceanography and Marine Life domains. The Platform for Ocean Knowledge Management (POKM) offers a services oriented framework to facilitate the sharing, discovery and visualization of multi-modal data and scientific models. To establish interoperability between two diverse domain, we have developed a common OWL-based domain ontology that captures and interrelates concepts from the two different domain. POKM also provide semantic descriptions of the functionalities of a range of e-research oriented web services through a OWL-S service ontology that supports dynamic discovery and invocation of services. POKM has been deployed as a web-based prototype system that is capable of fetching, sharing and visualizing marine animal detection and oceanographic data from multiple global data sources. 1 INTRODUCTION (POKM)—that offers a suite of knowledge-centric services for oceanographic researchers to (a) access, To comprehensively understand how changes to the share, integrate and operationalize the data, models ecosystem impact the ocean’s physical and and knowledge resources available at multiple sites; biological parameters (Cummings, 2005) (b) collaborate in joint scientific research oceanographers and marine biologists—termed as experiments by sharing resources, results, expertise the oceanographic research community—are seeking and models; and (c) form a virtual community of more collaboration in terms of sharing domain- researchers, marine resource managers, policy specific data and knowledge (Bos, 2007). -

An Application to Biology
Physical Science & Biophysics Journal ISSN: 2641-9165 Information as Order Hidden within Chance: An Application to Biology Strumia A* Review Article Istituto Nazionale di Alta Matematica "Francesco Severi", Italy Volume 3 Issue 3 *Corresponding author: Alberto Strumia, Istituto Nazionale di Alta Matematica Received Date: August 12, 2019 Published Date: August 27, 2019 "Francesco Severi", Italy, Email: [email protected] Abstract We show, by didactical examples, how algorithmic information (coded e.g., into a computer program) is required to build the structure of an organized system (either simple or complex). Ordered structures can be obtained as attractors both by some dynamics starting from sequential initial conditions (order from order) and by some dynamics starting from random initial conditions (order from chance) provided that a leading algorithmic information is assigned to govern the evolution of the generating process. In absence of information emergence of some ordered structure, like e.g., an organ of a living system is so highly improbable to be impossible in practice. We provide didactical examples of static models of a human heart, each generated starting either from ordered initial conditions, or from random sparse initial conditions, or more realistically by random cellular automata (so that any mother cell is allowed to generate a daughter cell only in a random contiguous location). Significantly, as it was pointed out by Gregory Chaitin, not all algorithmic information can be compressed into a string shorter than the sequence of its original individual code digits (incompressible information string). A question is still open about the DNA and, more generally, any biological information: is it to be considered as a compressible or an incompressible code string? In our example of anatomic human heart model we have treated the sequence of the co-ordinates of each sphere (roughly modeling a cell) as an uncompressed string, while a compressed program string seems to be able to provide only less realistic models.