Blueprint Genetics Cutis Laxa Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mackenzie's Mission Gene & Condition List
Mackenzie’s Mission Gene & Condition List What conditions are being screened for in Mackenzie’s Mission? Genetic carrier screening offered through this research study has been carefully developed. It is focused on providing people with information about their chance of having children with a severe genetic condition occurring in childhood. The screening is designed to provide genetic information that is relevant and useful, and to minimise uncertain and unclear information. How the conditions and genes are selected The Mackenzie’s Mission reproductive genetic carrier screen currently includes approximately 1300 genes which are associated with about 750 conditions. The reason there are fewer conditions than genes is that some genetic conditions can be caused by changes in more than one gene. The gene list is reviewed regularly. To select the conditions and genes to be screened, a committee comprised of experts in genetics and screening was established including: clinical geneticists, genetic scientists, a genetic pathologist, genetic counsellors, an ethicist and a parent of a child with a genetic condition. The following criteria were developed and are used to select the genes to be included: • Screening the gene is technically possible using currently available technology • The gene is known to cause a genetic condition • The condition affects people in childhood • The condition has a serious impact on a person’s quality of life and/or is life-limiting o For many of the conditions there is no treatment or the treatment is very burdensome for the child and their family. For some conditions very early diagnosis and treatment can make a difference for the child. -
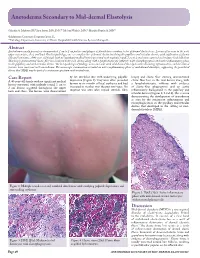
Anetoderma Secondary to Mid-Dermal Elastolysis
Anetoderma Secondary to Mid-dermal Elastolysis Gabriela A. Maloney, BS,* Jane James, MD, PhD,** Michael Welsch, MD,** Marylee Braniecki, MD** *Midwestern University, Downers Grove, IL **Pathology Department, University of Illinois Hospital & Health Sciences System, Chicago, IL Abstract Anetoderma usually presents as circumscribed, 1 cm to 2 cm patches and plaques of flaccid skin secondary to loss of dermal elastic tissue. Lesions often occur in the neck, upper extremities, chest, and back. On histopathology, one sees complete loss of dermal elastin involving the papillary and reticular dermis, with infiltration of plasma cells and histiocytes. A 40-year-old female with no significant medical history presented with multiple round, 1 cm to 2 cm lesions scattered on her upper back and chest. Skin biopsy demonstrated elastic-fiber loss localized to the mid-dermis along with a lymphohistiocytic infiltrate with elastophagocytosis and active inflammatory phase in the papillary and mid-reticular dermis. The histopathological findings were consistent with mid-dermal elastolysis with advancing inflammation, and the clinical features were consistent with anetoderma. The microscopic examination revealed an active inflammatory phase of mid-dermal elastolysis, supporting the postulated theory that MDE may be part of a continuous spectrum with anetoderma. Case Report by lax, wrinkled skin with underlying palpable biopsy and elastic-fiber staining demonstrated A 40-year-old female with no significant medical depression (Figure 1). They were often preceded elastic-fiber loss in the mid-dermis along with history presented with multiple round, 1 cm to by two to six months of local erythema and had a lymphohistiocytic infiltrate with evidence 2 cm lesions scattered throughout the upper increased in number over the past two years. -

Centometabolic® COMBINING GENETIC and BIOCHEMICAL TESTING for the FAST and COMPREHENSIVE DIAGNOSTIC of METABOLIC DISORDERS
CentoMetabolic® COMBINING GENETIC AND BIOCHEMICAL TESTING FOR THE FAST AND COMPREHENSIVE DIAGNOSTIC OF METABOLIC DISORDERS GENE ASSOCIATED DISEASE(S) ABCA1 HDL deficiency, familial, 1; Tangier disease ABCB4 Cholestasis, progressive familial intrahepatic 3 ABCC2 Dubin-Johnson syndrome ABCD1 Adrenoleukodystrophy; Adrenomyeloneuropathy, adult ABCD4 Methylmalonic aciduria and homocystinuria, cblJ type ABCG5 Sitosterolemia 2 ABCG8 Sitosterolemia 1 ACAT1 Alpha-methylacetoacetic aciduria ADA Adenosine deaminase deficiency AGA Aspartylglucosaminuria AGL Glycogen storage disease IIIa; Glycogen storage disease IIIb AGPS Rhizomelic chondrodysplasia punctata, type 3 AGXT Hyperoxaluria, primary, type 1 ALAD Porphyria, acute hepatic ALAS2 Anemia, sideroblastic, 1; Protoporphyria, erythropoietic, X-linked ALDH4A1 Hyperprolinemia, type II ALDOA Glycogen storage disease XII ALDOB Fructose intolerance, hereditary ALG3 Congenital disorder of glycosylation, type Id ALPL Hypophosphatasia, adult; Hypophosphatasia, childhood; Hypophosphatasia, infantile; Odontohypophosphatasia ANTXR2 Hyaline fibromatosis syndrome APOA2 Hypercholesterolemia, familial, modifier of APOA5 Hyperchylomicronemia, late-onset APOB Hypercholesterolemia, familial, 2 APOC2 Hyperlipoproteinemia, type Ib APOE Sea-blue histiocyte disease; Hyperlipoproteinemia, type III ARG1 Argininemia ARSA Metachromatic leukodystrophy ARSB Mucopolysaccharidosis type VI (Maroteaux-Lamy) 1 GENE ASSOCIATED DISEASE(S) ASAH1 Farber lipogranulomatosis; Spinal muscular atrophy with progressive myoclonic epilepsy -

De Barsy Syndrome
orphananesthesia Anesthesia recommendations for patients suffering from De Barsy syndrome Disease name: De Barsy syndrome ICD 10: Q87.7; OMIM 614438 Synonyms: DBS, De Barsy-Moens-Dierckx syndrome, Progeroid syndrome of De Barsy, Autosomal recessive cutis laxa Type 3 *With 2 gene subdivisions: ARCL3A: caused by a ALDH18A1 mutation ARCL3B: caused by a PYCR1 mutation DeBarsy syndrome is a rare clinical syndrome characterized by cutis laxa, ophthalmic opacification, skeletal malformations, as well as mental and growth retardation. This disease is genetically transmitted in an autosomal recessive fashion. Affected patients often require surgical correction of ophthalmic and orthopaedic abnormalities. This syndrome was first described by A.M. De Barsy in 1967 and less than 100 known cases are documented in the medical literature. Very little has been published on this rare disorder and only a single article has addressed anesthesia case outcomes and management strategies (Aponte, 2010). The diverse collection of clinical manifestations in De Barsy syndrome includes: intra-uterine growth retardation (IUGR), postnatal growth delay, motor delay, cognitive impairment, hypontonia, athetoid movements, malformations, microcephaly, wormian bones, large fontanelles, facial dysmorphism, cataracts, corneal clouding, thin/wrinkled skin, easy bruising, sparse hair, joint laxity, osteopenia, and inguinal hernias. Medicine in progress Perhaps new knowledge Every patient is unique Perhaps the diagnostic is wrong Find more information on the disease, its centres -

Dermatologists Cannot Be Lax About Acquired Cutis Laxa by Warren R
Dermatologists cannot be lax about acquired cutis laxa By Warren R. Heymann, MD Jan. 28, 2019 Acral localized acquired cutis laxa. Loose, redundant skin on the volar finger pads bilaterally that gave them a flat, rounded appearance. Credit: JAADFrom the time I was a medical student, I have always been fascinated by the “little old man” appearance of patient with cutis laxa (CL). In their excellent review, Berk et al note that CL is characterized by abnormal elastic fibers resulting in loose, redundant skin. Typically, skin in CL can easily be pulled and only slowly returns to its original position. Unlike some conditions in the differential diagnosis, notably Ehlers-Danlos syndrome(s) or pseudoxanthoma elasticum, easy bruising or abnormal scarring does not characterize CL. Redundant skin is often most noticeable on the neck, hands, and groin, but can also be seen on the face, creating a premature aging appearance. CL may be inherited or acquired (ACL). Inherited forms include autosomal dominant CL; autosomal recessive CL (ARCL)-I, -IIA, and -IIB; X-linked CL and other variants. Although all of the inherited forms of CL are rare, ARCL has most commonly been reported, particularly ARCL- II. Because of significant overlap among these types, precise clinical classification can be difficult. For the clinical differentiation of the variants and their genetic mutations, please refer to Berk et al (1). The etiology of CL involves abnormalities the elastic fiber network in skin and internal organs. Autosomal dominant CL has been associated with mutations of the elastin gene. X-linked CL is associated with gene mutations ATPA7 and abnormalities of copper transport. -

Pili Torti: a Feature of Numerous Congenital and Acquired Conditions
Journal of Clinical Medicine Review Pili Torti: A Feature of Numerous Congenital and Acquired Conditions Aleksandra Hoffmann 1 , Anna Wa´skiel-Burnat 1,*, Jakub Z˙ ółkiewicz 1 , Leszek Blicharz 1, Adriana Rakowska 1, Mohamad Goldust 2 , Małgorzata Olszewska 1 and Lidia Rudnicka 1 1 Department of Dermatology, Medical University of Warsaw, Koszykowa 82A, 02-008 Warsaw, Poland; [email protected] (A.H.); [email protected] (J.Z.);˙ [email protected] (L.B.); [email protected] (A.R.); [email protected] (M.O.); [email protected] (L.R.) 2 Department of Dermatology, University Medical Center of the Johannes Gutenberg University, 55122 Mainz, Germany; [email protected] * Correspondence: [email protected]; Tel.: +48-22-5021-324; Fax: +48-22-824-2200 Abstract: Pili torti is a rare condition characterized by the presence of the hair shaft, which is flattened at irregular intervals and twisted 180◦ along its long axis. It is a form of hair shaft disorder with increased fragility. The condition is classified into inherited and acquired. Inherited forms may be either isolated or associated with numerous genetic diseases or syndromes (e.g., Menkes disease, Björnstad syndrome, Netherton syndrome, and Bazex-Dupré-Christol syndrome). Moreover, pili torti may be a feature of various ectodermal dysplasias (such as Rapp-Hodgkin syndrome and Ankyloblepharon-ectodermal defects-cleft lip/palate syndrome). Acquired pili torti was described in numerous forms of alopecia (e.g., lichen planopilaris, discoid lupus erythematosus, dissecting Citation: Hoffmann, A.; cellulitis, folliculitis decalvans, alopecia areata) as well as neoplastic and systemic diseases (such Wa´skiel-Burnat,A.; Zółkiewicz,˙ J.; as cutaneous T-cell lymphoma, scalp metastasis of breast cancer, anorexia nervosa, malnutrition, Blicharz, L.; Rakowska, A.; Goldust, M.; Olszewska, M.; Rudnicka, L. -
The Skin in Genetically-Controlled Metabolic Disorders P
Review Article J Med Genet: first published as 10.1136/jmg.10.2.101 on 1 June 1973. Downloaded from Journal of Medical Genetics (1973). 10, 101. The Skin in Genetically-controlled Metabolic Disorders P. C. H. NEWBOLD Department of Medicine, Cambridge University Medical School, Hills Road, Cambridge CB2 2QL Diseased nature oftentimes breaks forth however, it may lead to difficulty in assessing intelli- In strange eruptions.-Henry IV, part 1, III, i. gence, which is within normal range in 50% of The skin is now commonly accepted as a mirror patients. There is suggestive evidence of a re- of internal disease, but as with other looking-glasses, lationship between subnormal folate levels and low the evidence offered may be selected or ignored. intelligence (Carey et al, 1968), and studies have The epidermis is an interesting structure, especially shown increased turnover of folate coenzymes and rich in tyrosine, phenylalanine, tryptophan, and resulting folate depletion in these patients (Carey et histidine, when compared with the corium (Roth- al, 1968; Butterworth, Krumdieck, and Baugh, man, 1965). Tyrosinaemia and histidinaemia do 1971). There is also a high incidence of an organic not include cutaneous manifestations, but culture of brain syndrome following intracranial vascular skin fibroblasts is a helpful diagnostic tool for study- thromboses (Dunn, Perry, and Dolman, 1966), ing metabolic defects such as citrullinaemia, cysti- copyright. nosis, and maple-syrup urine disease (Scriver, 1969). Most of the conditions now to be described are rare, but if these metabolic diseases were com- mon, there could be no human race as we know it. Homocystinuria http://jmg.bmj.com/ This most informative anomaly was discovered during a study of mentally retarded patients in Ire- land (Carson and Neill, 1962). -

Avero® Exon Global 280+ Genes
Avero® Exon Global 280+ genes CARRIER DETECTION RESIDUAL GENE DISORDER NAME ETHNICITY FREQUENCY RATE RISK ABCB11 Progressive familial intrahepatic cholestasis, type II General Population 1 in 158 99% 1 in 15701 ABCC8 Familial hyperinsulinism, ABCC8-related General Population 1 in 112 99% 1 in 11101 Ashkenazi Jewish 1 in 52 99% 1 in 5101 Finnish 1 in 29 99% 1 in 2801 ABCD1 Adrenoleukodystrophy, X-linked General Population 1 in 10500 99% 1 in 1049901 Sephardic Jewish 1 in 10500 99% 1 in 1049901 ACAD9 Riboflavin responsive complex 1 deficiency (acyl-coenzyme General Population <1 in 500 99% 1 in 49901 dehydrogenase 9 deficiency) ACADM Medium-chain acyl-CoA dehydrogenase deficiency General Population 1 in 35 99% 1 in 3401 Asian 1 in 178 99% 1 in 17701 Caucasian 1 in 64 99% 1 in 6300 ACADVL Very long-chain acyl-CoA dehydrogenase deficiency General Population 1 in 86 99% 1 in 8500 Asian 1 in 194 99% 1 in 19301 Caucasian 1 in 88 99% 1 in 8700 ACAT1 Beta-ketothiolase deficiency General Population 1 in 347 99% 1 in 34601 Asian 1 in 289 99% 1 in 28801 Caucasian 1 in 354 99% 1 in 35301 ACOX1 Acyl-CoA oxidase I deficiency General Population <1 in 500 99% 1 in 49901 ACSF3 Combined malonic and methylmalonic aciduria General Population 1 in 86 99% 1 in 8500 ADA Adenosine deaminase deficiency General Population 1 in 224 99% 1 in 22301 ADAMTS2 Ehlers-Danlos syndrome, type VIIC General Population <1 in 500 99% 1 in 49901 Ashkenazi Jewish 1 in 248 99% 1 in 24701 6221 Riverside Drive, Suite 119, Irving, TX 75039 I Tel 877.232.9924 I averodx.com ©2020 Avero Diagnostics. -

RIN2 Deficiency Results in Macrocephaly, Alopecia, Cutis Laxa
REPORT RIN2 Deficiency Results in Macrocephaly, Alopecia, Cutis Laxa, and Scoliosis: MACS Syndrome Lina Basel-Vanagaite,1,2,14 Ofer Sarig,4,14 Dov Hershkovitz,6,7 Dana Fuchs-Telem,2,4 Debora Rapaport,3 Andrea Gat,5 Gila Isman,4 Idit Shirazi,4 Mordechai Shohat,1,2 Claes D. Enk,10 Efrat Birk,2 Ju¨rgen Kohlhase,11 Uta Matysiak-Scholze,11 Idit Maya,1 Carlos Knopf,9 Anette Peffekoven,12 Hans-Christian Hennies,12 Reuven Bergman,8 Mia Horowitz,3 Akemi Ishida-Yamamoto,13 and Eli Sprecher2,4,6,* Inherited disorders of elastic tissue represent a complex and heterogeneous group of diseases, characterized often by sagging skin and occasionally by life-threatening visceral complications. In the present study, we report on an autosomal-recessive disorder that we have termed MACS syndrome (macrocephaly, alopecia, cutis laxa, and scoliosis). The disorder was mapped to chromosome 20p11.21-p11.23, and a homozygous frameshift mutation in RIN2 was found to segregate with the disease phenotype in a large consan- guineous kindred. The mutation identified results in decreased expression of RIN2, a ubiquitously expressed protein that interacts with Rab5 and is involved in the regulation of endocytic trafficking. RIN2 deficiency was found to be associated with paucity of dermal micro- fibrils and deficiency of fibulin-5, which may underlie the abnormal skin phenotype displayed by the patients. Disorders of elastic tissue often share common phenotypic shown to result in decreased secretion of elastin.9,10 In features, including redundant skin, hyperlaxity of joints, addition, -

Molecular Genetics and Metabolism Reports 24 (2020) 100625
Molecular Genetics and Metabolism Reports 24 (2020) 100625 Contents lists available at ScienceDirect Molecular Genetics and Metabolism Reports journal homepage: www.elsevier.com/locate/ymgmr Targeted next generation sequencing for newborn screening of Menkes T disease ⁎ Richard B. Parada,1, Stephen G. Kalerb,e, ,1, Evan Maucelic, Tanya Sokolskyc,d, Ling Yib, Arindam Bhattacharjeec,d a Department of Pediatric Newborn Medicine, Brigham & Women's Hospital, Harvard Medical School, Boston, MA, United States of America b Section on Translational Neuroscience, Molecular Medicine Branch, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD, United States of America c Parabase Genomics, Inc., Boston, MA, United States of America d Baebies, Inc., Durham, NC, United States of America e Center for Gene Therapy, Abigail Wexner Research Institute, Nationwide Children's Hospital, Columbus, OH, United States of America ARTICLE INFO ABSTRACT Keywords: Purpose: Population-based newborn screening (NBS) allows early detection and treatment of inherited disorders. Menkes disease For certain medically-actionable conditions, however, NBS is limited by the absence of reliable biochemical ATP7A signatures amenable to detection by current platforms. We sought to assess the analytic validity of an ATP7A Molecular genetics targeted next generation DNA sequencing assay as a potential newborn screen for one such disorder, Menkes Targeted next generation sequencing disease. Dried blood spots Methods: Dried blood spots from control or Menkes disease subjects (n = 22) were blindly analyzed for pa- Newborn screening thogenic variants in the copper transport gene, ATP7A. The analytical method was optimized to minimize cost and provide rapid turnaround time. Results: The algorithm correctly identified pathogenic ATP7A variants, including missense, nonsense, small insertions/deletions, and large copy number variants, in 21/22 (95.5%) of subjects, one of whom had incon- clusive diagnostic sequencing previously. -

Genetics of Lipedema: New Perspectives on Genetic Research and Molecular Diagnoses S
European Review for Medical and Pharmacological Sciences 2019; 23: 5581-5594 Genetics of lipedema: new perspectives on genetic research and molecular diagnoses S. PAOLACCI1, V. PRECONE2, F. ACQUAVIVA3, P. CHIURAZZI4,5, E. FULCHERI6,7, M. PINELLI3,8, F. BUFFELLI9,10, S. MICHELINI11, K.L. HERBST12, V. UNFER13, M. BERTELLI2; GENEOB PROJECT 1MAGI’S LAB, Rovereto (TN), Italy 2MAGI EUREGIO, Bolzano, Italy 3Department of Translational Medicine, Section of Pediatrics, Federico II University, Naples, Italy 4Istituto di Medicina Genomica, Fondazione A. Gemelli, Università Cattolica del Sacro Cuore, Rome, Italy 5UOC Genetica Medica, Fondazione Policlinico Universitario “A. Gemelli” IRCCS, Rome, Italy 6Fetal and Perinatal Pathology Unit, IRCCS Istituto Giannina Gaslini, Genoa, Italy 7Department of Integrated Surgical and Diagnostic Sciences, University of Genoa, Genoa, Italy 8Telethon Institute of Genetics and Medicine (TIGEM), Pozzuoli, Italy 9Fetal and Perinatal Pathology Unit, IRCCS Istituto Giannina Gaslini, Genoa, Italy 10Department of Neuroscience, Rehabilitation, Ophthalmology, Genetics and Maternal-Infantile Sciences, University of Genoa, Genoa, Italy 11Department of Vascular Rehabilitation, San Giovanni Battista Hospital, Rome, Italy 12Department of Medicine, University of Arizona, Tucson, AZ, USA 13Department of Developmental and Social Psychology, Faculty of Medicine and Psychology, Sapienza University of Rome, Rome, Italy Abstract. – OBJECTIVE: The aim of this quali- Introduction tative review is to provide an update on the cur- rent understanding of the genetic determinants of lipedema and to develop a genetic test to dif- Lipedema is an underdiagnosed chronic debil- ferentiate lipedema from other diagnoses. itating disease characterized by bruising and pain MATERIALS AND METHODS: An electronic and excess of subcutaneous adipose tissue of the search was conducted in MEDLINE, PubMed, and legs and/or arms in women during or after times Scopus for articles published in English up to of hormone change, especially in puberty1. -

Acquired Cutis Laxa of Face with Multiple Myeloma
Letters to the Editor 6. Takayama K, Satoh T, Hayashi M, Yokozeki H. Psoriatic skin Serum β2‑microglobulin was increased. Bone marrow lesions induced by BCG vaccination. Acta Derm Venereol examination revealed 14% plasma cells. Based on 2008;88:621‑2. 7. Monacelli M. Koebner reaction in psoriasis. In: Farber EM, these findings, a diagnosis of early multiple myeloma Cox AJ, editors. Psoriasis: Proceedings of the International with nephrotic syndrome was made. She was started Symposium. Vol. 99. Stanford: Stanford University press; 1971. p. 104‑11. on a chemotherapy regimen consisting of thalidomide 8. Rambukkana A, Das PK, Witkamp L, Yong S, Meinardi MM, (100 mg/day), oral cyclophosphamide (150 mg/day) Bos JD. Antibodies to mycobacterial 65‑kDa heat shock protein both for the first five days in every month with weekly and other immunodominantantigens in patients with psoriasis. J Invest Dermatol 1993;100:87‑92. intravenous dexamethasone (40 mg/week) along with loop diuretics. In addition, the patient was prescribed Access this article online bortezomib which was discontinued by the patient after two months. Following one year of treatment, the Quick Response Code: Website: www.ijdvl.com anasarca subsided leaving behind loose wrinkled skin all over the body. Over the next one year, the skin over the DOI: 10.4103/0378-6323.140309 extremities and the abdomen reverted back to normal; however, loosening of skin persisted over the face. PMID: ***** Examination revealed a healthy‑appearing woman, who appeared older than her stated age [Figure 1]. She Acquired cutis laxa of face with had loose, sagging skin on the face and neck giving her a hound‑dog facies [Figure 2].