Lung Microbiome Participation in Local Immune Response Regulation in Respiratory Diseases
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
IFM Innate Immunity Infographic
UNDERSTANDING INNATE IMMUNITY INTRODUCTION The immune system is comprised of two arms that work together to protect the body – the innate and adaptive immune systems. INNATE ADAPTIVE γδ T Cell Dendritic B Cell Cell Macrophage Antibodies Natural Killer Lymphocites Neutrophil T Cell CD4+ CD8+ T Cell T Cell TIME 6 hours 12 hours 1 week INNATE IMMUNITY ADAPTIVE IMMUNITY Innate immunity is the body’s first The adaptive, or acquired, immune line of immunological response system is activated when the innate and reacts quickly to anything that immune system is not able to fully should not be present. address a threat, but responses are slow, taking up to a week to fully respond. Pathogen evades the innate Dendritic immune system T Cell Cell Through antigen Pathogen presentation, the dendritic cell informs T cells of the pathogen, which informs Macrophage B cells B Cell B cells create antibodies against the pathogen Macrophages engulf and destroy Antibodies label invading pathogens pathogens for destruction Scientists estimate innate immunity comprises approximately: The adaptive immune system develops of the immune memory of pathogen exposures, so that 80% system B and T cells can respond quickly to eliminate repeat invaders. IMMUNE SYSTEM AND DISEASE If the immune system consistently under-responds or over-responds, serious diseases can result. CANCER INFLAMMATION Innate system is TOO ACTIVE Innate system NOT ACTIVE ENOUGH Cancers grow and spread when tumor Certain diseases trigger the innate cells evade detection by the immune immune system to unnecessarily system. The innate immune system is respond and cause excessive inflammation. responsible for detecting cancer cells and This type of chronic inflammation is signaling to the adaptive immune system associated with autoimmune and for the destruction of the cancer cells. -

Lung Anatomy
Lung anatomy Breathing Breathing is an automatic and usually subconscious process which is controlled by the brain. The brain will determine how much oxygen we require and how fast we need to breathe in order to supply our vital organs (brain, heart, kidneys, liver, stomach and bowel), as well as our muscles and joints, with enough oxygen to carry out our normal daily activities. In order for breathing to be effective we need to use our lungs, breathing muscles and blood system efficiently. This leaflet should help you to better understand the process of breathing and how we get the much needed oxygen into our bodies. The lungs You have two lungs, one in the right side and one in the left side of your chest. The right lung is bigger than the left due to the position of the heart (which is positioned in the left side of the chest). Source: Pulmonary Rehabilitation Reference No: 66354-1 Issue date: 13/2/20 Review date: 13/2/23 Page 1 of 5 Both lungs are covered by 2 thin layers of tissue called the pleura. The pleura stop the surface of the lungs rubbing together as we breathe in and out. The lungs are protected by the ribcage. The airways Within the lungs there is a vast network of airways (tubes) which help to transport the oxygen into the lungs and the carbon dioxide out. These tubes branch into smaller and smaller tubes the further they go into the lungs. Page 2 of 5 Trachea (windpipe): This tube connects your nose and mouth to your lungs. -

Our Immune System (Children's Book)
OurOur ImmuneImmune SystemSystem A story for children with primary immunodeficiency diseases Written by IMMUNE DEFICIENCY Sara LeBien FOUNDATION A note from the author The purpose of this book is to help young children who are immune deficient to better understand their immune system. What is a “B-cell,” a “T-cell,” an “immunoglobulin” or “IgG”? They hear doctors use these words, but what do they mean? With cheerful illustrations, Our Immune System explains how a normal immune system works and what treatments may be necessary when the system is deficient. In this second edition, a description of a new treatment has been included. I hope this book will enable these children and their families to explore together the immune system, and that it will help alleviate any confusion or fears they may have. Sara LeBien This book contains general medical information which cannot be applied safely to any individual case. Medical knowledge and practice can change rapidly. Therefore, this book should not be used as a substitute for professional medical advice. SECOND EDITION COPYRIGHT 1990, 2007 IMMUNE DEFICIENCY FOUNDATION Copyright 2007 by Immune Deficiency Foundation, USA. Readers may redistribute this article to other individuals for non-commercial use, provided that the text, html codes, and this notice remain intact and unaltered in any way. Our Immune System may not be resold, reprinted or redistributed for compensation of any kind without prior written permission from Immune Deficiency Foundation. If you have any questions about permission, please contact: Immune Deficiency Foundation, 40 West Chesapeake Avenue, Suite 308, Towson, MD 21204, USA; or by telephone at 1-800-296-4433. -

Cells, Tissues and Organs of the Immune System
Immune Cells and Organs Bonnie Hylander, Ph.D. Aug 29, 2014 Dept of Immunology [email protected] Immune system Purpose/function? • First line of defense= epithelial integrity= skin, mucosal surfaces • Defense against pathogens – Inside cells= kill the infected cell (Viruses) – Systemic= kill- Bacteria, Fungi, Parasites • Two phases of response – Handle the acute infection, keep it from spreading – Prevent future infections We didn’t know…. • What triggers innate immunity- • What mediates communication between innate and adaptive immunity- Bruce A. Beutler Jules A. Hoffmann Ralph M. Steinman Jules A. Hoffmann Bruce A. Beutler Ralph M. Steinman 1996 (fruit flies) 1998 (mice) 1973 Discovered receptor proteins that can Discovered dendritic recognize bacteria and other microorganisms cells “the conductors of as they enter the body, and activate the first the immune system”. line of defense in the immune system, known DC’s activate T-cells as innate immunity. The Immune System “Although the lymphoid system consists of various separate tissues and organs, it functions as a single entity. This is mainly because its principal cellular constituents, lymphocytes, are intrinsically mobile and continuously recirculate in large number between the blood and the lymph by way of the secondary lymphoid tissues… where antigens and antigen-presenting cells are selectively localized.” -Masayuki, Nat Rev Immuno. May 2004 Not all who wander are lost….. Tolkien Lord of the Rings …..some are searching Overview of the Immune System Immune System • Cells – Innate response- several cell types – Adaptive (specific) response- lymphocytes • Organs – Primary where lymphocytes develop/mature – Secondary where mature lymphocytes and antigen presenting cells interact to initiate a specific immune response • Circulatory system- blood • Lymphatic system- lymph Cells= Leukocytes= white blood cells Plasma- with anticoagulant Granulocytes Serum- after coagulation 1. -

Hormones and the Immune Response
Ann Rheum Dis: first published as 10.1136/ard.48.1.1 on 1 January 1989. Downloaded from Annals of the Rheumatic Diseases, 1989; 48, 1-6 Review Hormones and the immune response Recent advances suggest that the immune system cells, are present on mouse spleen cells3 and human does not function in isolation but is influenced by peripheral blood mononuclear cells.4 Receptors, other physiological systems such as the endocrine identical to those in the central nervous system, for and neuroendocrine systems. This review discusses methionine enkephalin are present on splenocytes aspects of immune function altered by neuroendo- and T lymphocytes.3 In contrast, leucine enkephalin crine peptides, sex hormones, and vitamin D and j3-endorphin receptors on T lymphocyte differ metabolites. from those in the central nervous system as binding cannot be inhibited by opiate antagonists.5 6 In the Neuroendocrine effects case of ,3-endorphin the bindings occur through its carboxy terminal, whereas opiates bind their A system of bidirectional communication between receptor through the amino terminus. This raises the immune and neuroendocrine system exists, in an interesting possibility that a peptide such as which the two systems share a common set of f6-endorphin could form a bridge between two hormones and receptors.'2 Not only do immune lymphocyte subtypes by binding to one through its for peptides, to the and cells possess receptors neuroendocrine amino terminus opiate receptor through copyright. they are also capable of synthesising them and of its carboxy terminus to the non-opiate receptor on responding to them. Products of immune cells affect another lymphocyte.4 the central nervous system, which possesses recep- Other neuroendocrine peptide receptors present tors for cytokines and can also synthesise them on leucocztes include those for neurotensin,7 sub- (Fig. -

Manatee Anatomy and Physiology
Manatee Anatomy and Physiology Grade level: Elementary 5 Subject Area: Biology, Anatomy and Physiology, Marine Biology Duration: Teach: 15 minutes, Activity: 20 minutes, Discussion: 20 minutes. Setting: Classroom Next Generation Sunshine State Standards: Reading: LAFS.5.RI Reading standards for Information text Cluster 3 LAFS.5.RI.3 Writing: LA.B.2.1, LA.B.1.2 Speaking and Listening: Cluster 2 LAFS.5.SL.Z Presentation of Knowledge and Ideas Science: Body of Knowledge Life Science: SC.5.L.14, SC.5.L.15, SC.5.L.17, Body of Knowledge Nature of Science Supporting Idea 1: SC.5.N.1 Practice of Science, Supporting Idea 2:SC.5.N.2 Characteristics of Scientific Knowledge Earth Systems and Patterns: 7SC.E.E.7 Statewide Science Assessment Prompt: How might humans help manatees survive? Objectives: Students will learn about manatee bodies and explain some anatomical and physiological differences between manatees, humans and other animals. Materials: Handouts of basic manatee anatomy, dolphin anatomy & human anatomy, crayons or markers, coloring direction sheet, question worksheet, Quiz sheet Vocabulary: Mammal, endangered species, habitat, conservation, vibrissae, nares, blowhole, flipper, herbivore, omnivore, carnivore. Background/Preparation: Handouts of manatee, dolphin, and human anatomy. Fact sheets comparing and contrasting specific and unique anatomical aspects of each species. Basic manatee fact sheet highlighting personality, limited habitat, endangered status and conservation efforts. Teachers can review the manatee fact sheets, and select points of interest they would most like to incorporate into a lesson. This activity may fit best into the week where the human anatomy lessons are addressed. Teachers can present the information via traditional lecture, group discussion, question and answer session, or doing the coloring activity as the lesson points are addressed, etc. -

The Immune System
Chapter 43 The Immune System Lecture Outline Overview: Reconnaissance, Recognition, and Response • An animal must defend itself against pathogens, infectious agents that cause disease. o Viruses, bacteria, protists, and fungi infect a wide range of animals, including humans. • Animals fight back in various ways. o Immune cells patrol the body fluids of animals, seeking out and destroying foreign cells. o Responses to infection include proteins that punch holes in bacterial membranes or block viruses from entering body cells. • Immune systems help animals to avoid or limit many infections. o External barriers, formed by the skin or shell, provide an obstacle to microbes. o Chemical secretions that trap or kill microbes guard the body’s entrances and exits. o The internal defenses include macrophages and other phagocytic cells that ingest and destroy pathogens. • An animal’s immune system must detect foreign particles and tissues that invade the body, distinguishing self from nonself. • To identify pathogens, animal immune systems use receptors that specifically bind molecules from foreign cells or viruses. • Two general strategies for molecular recognition form the basis for innate and acquired immunity. • Innate immunity is common to all animals. • Innate immune responses are active immediately upon infection and are the same whether or not the pathogen has been encountered previously. • Innate immunity includes the barrier defenses (for example, skin) as well as defenses that combat pathogens after they enter the body. • The activation of many of these internal defenses relies on the recognition of pathogens. o Innate immune cells produce a small, preset group of receptor proteins that accomplish this recognition. -

The Fifth Pulmonary Vein
CASE REPORT Anatomy Journal of Africa. 2016. Vol 5 (2): 704 - 706 The Fifth Pulmonary vein Hilkiah Kinfemichael, Asrat Dawit Correspondence to Hilkiah Kinfemichael, Myungsung Medical College, Addis Ababa, Ethiopia PO Box 14972 Email : [email protected]. ABSTRACT A cadaver in Myungsung Medical College (MMC) had a 3rd pulmonary vein originating from the middle lobe of the right lung. Such anatomical variations are very rare. People with this variation have a total of five pulmonary veins entering left atrium. It has clinical implications especially for thoracic surgeons and radiologists during radiofrequency ablations, lobectomies, valve replacements, pulmonary vein catheterizations, video-assisted thoracic surgery (VATS) and others. Key Words: Anatomy, Variations, Pulmonary veins. INTRODUCTION Two pulmonary veins usually drain oxygenated upper lobe, is formed by the union of blood from each lung to left atrium of the apicoposterior, anterior and lingular veins. The heart. The lobular tributaries lie mainly in the inferior left pulmonary vein, which drains the interlobular septa and two pulmonary veins lower lobe, is formed by the hilar union of two from each lung enter left atrium through two veins, superior and common basal veins. The separate pulmonary ostia on either side. On the right and left pulmonary veins perforate the right lung veins from the apical, anterior and fibrous pericardium and open separately in the posterior part of upper lobe unite with a middle posterosuperior aspect of the left atrium lobar vein, which is formed by lateral and (Standring et al., 2008). This anatomy could medial tributaries, and form the superior right show variations, with more veins draining pulmonary vein (Standring, 2008). -
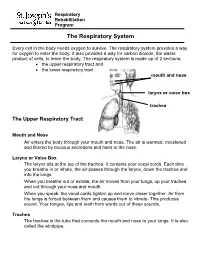
The Respiratory System
Respiratory Rehabilitation Program The Respiratory System Every cell in the body needs oxygen to survive. The respiratory system provides a way for oxygen to enter the body. It also provides a way for carbon dioxide, the waste product of cells, to leave the body. The respiratory system is made up of 2 sections: the upper respiratory tract and the lower respiratory tract mouth and nose larynx or voice box trachea The Upper Respiratory Tract Mouth and Nose Air enters the body through your mouth and nose. The air is warmed, moistened and filtered by mucous secretions and hairs in the nose. Larynx or Voice Box The larynx sits at the top of the trachea. It contains your vocal cords. Each time you breathe in or inhale, the air passes through the larynx, down the trachea and into the lungs. When you breathe out or exhale, the air moves from your lungs, up your trachea and out through your nose and mouth. When you speak, the vocal cords tighten up and move closer together. Air from the lungs is forced between them and causes them to vibrate. This produces sound. Your tongue, lips and teeth form words out of these sounds. Trachea The trachea is the tube that connects the mouth and nose to your lungs. It is also called the windpipe. The Lower Respiratory Tract Inside Lungs Outside Lungs bronchial tubes alveoli diaphragm (muscle) Bronchial Tubes The trachea splits into 2 bronchial tubes in your lungs. These are called the left bronchus and right bronchus. The bronchus tubes keep branching off into smaller and smaller tubes called bronchi. -

Understanding the Immune System: How It Works
Understanding the Immune System How It Works U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES NATIONAL INSTITUTES OF HEALTH National Institute of Allergy and Infectious Diseases National Cancer Institute Understanding the Immune System How It Works U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES NATIONAL INSTITUTES OF HEALTH National Institute of Allergy and Infectious Diseases National Cancer Institute NIH Publication No. 03-5423 September 2003 www.niaid.nih.gov www.nci.nih.gov Contents 1 Introduction 2 Self and Nonself 3 The Structure of the Immune System 7 Immune Cells and Their Products 19 Mounting an Immune Response 24 Immunity: Natural and Acquired 28 Disorders of the Immune System 34 Immunology and Transplants 36 Immunity and Cancer 39 The Immune System and the Nervous System 40 Frontiers in Immunology 45 Summary 47 Glossary Introduction he immune system is a network of Tcells, tissues*, and organs that work together to defend the body against attacks by “foreign” invaders. These are primarily microbes (germs)—tiny, infection-causing Bacteria: organisms such as bacteria, viruses, streptococci parasites, and fungi. Because the human body provides an ideal environment for many microbes, they try to break in. It is the immune system’s job to keep them out or, failing that, to seek out and destroy them. Virus: When the immune system hits the wrong herpes virus target or is crippled, however, it can unleash a torrent of diseases, including allergy, arthritis, or AIDS. The immune system is amazingly complex. It can recognize and remember millions of Parasite: different enemies, and it can produce schistosome secretions and cells to match up with and wipe out each one of them. -

Replication in the Lung and Immune System Alerted
Replication in the lung and immune system alerted The viral load study in Germany showed that active viral replication occurs in the upper respiratory tract. Seven out of nine participants listed a cough among their initial symptoms. In contrast to the falling numbers of viral units in the upper respiratory tract, numbers in sputum rose for most of the participants. In two individuals with some signs of lung infection, the virus in sputum peaked at day 10- 11. It was present in the sputum up to day 28 in one person. Across all participants, there was an average of 7 million units millilitre (about 35 million units a teaspoon), this amount is about 1000 times more than that in people with SARS In the lung, the ACE2 receptor sits on top of lung cells called pneumocystis. These have an important role in producing surfactant- a compound that coats the air sacs (alveoli), thus helping maintain enough surface tension to keep the sacs open for the exchange of oxygen and carbon dioxide. As soon as the body recognizes foreign protein, it mounts the first response. One part of the body’s immune response- The lymphocytes- begin to produce the first defence IgM-type antibodies and then the longer-term specific neutralizing antibodies (the IgG type). In the German viral study, 50% of the participants had IgM or IgG antibodies by day 7, and they all had these antibodies by day 14. The amount of antibodies did not predict the clinical course of the disease. 80% of people with COVID-19 will have mild or asymptomatic disease, with common symptoms including fever, cough, and loss of sense of smell. -

Lung Lobe Segmentation Based on Lung Fissure Surface Classification
algorithms Article Lung Lobe Segmentation Based on Lung Fissure Surface Classification Using a Point Cloud Region Growing Approach Xin Chen 1, Hong Zhao 2 and Ping Zhou 3,* 1 College of Electrical and Information Engineering, Hunan University, Changsha 410082, China; [email protected] 2 College of Aerospace Science and Engineering, National University of Defense Technology, Changsha 410003, China; [email protected] 3 College of Biology, Hunan University, Changsha 410082, China * Correspondence: [email protected] Received: 2 September 2020; Accepted: 13 October 2020; Published: 15 October 2020 Abstract: In anatomy, the lung can be divided by lung fissures into several pulmonary lobe units with specific functions. Identifying the lung lobes and the distribution of various diseases among different lung lobes from CT images is important for disease diagnosis and tracking after recovery. In order to solve the problems of low tubular structure segmentation accuracy and long algorithm time in segmenting lung lobes based on lung anatomical structure information, we propose a segmentation algorithm based on lung fissure surface classification using a point cloud region growing approach. We cluster the pulmonary fissures, transformed into point cloud data, according to the differences in the pulmonary fissure surface normal vector and curvature estimated by principal component analysis. Then, a multistage spline surface fitting method is used to fill and expand the lung fissure surface to realize the lung lobe segmentation. The proposed approach was qualitatively and quantitatively evaluated on a public dataset from Lobe and Lung Analysis 2011 (LOLA11), and obtained an overall score of 0.84. Although our approach achieved a slightly lower overall score compared to the deep learning based methods (LobeNet_V2 and V-net), the inter-lobe boundaries from our approach were more accurate for the CT images with visible lung fissures.