Helix-Loop-Helix Proteins Regulate Pre-TCR and TCR Signaling Through Modulation of Rel/NF- B Activities
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Novel TAL1 Targets Beyond Protein-Coding Genes: Identification of TAL1-Regulated Micrornas in T-Cell Acute Lymphoblastic Leukemia
Letters to the Editor 1603 REFERENCES 8 Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R et al. Frequent 1 Rozman C, Montserrat E. Chronic lymphocytic leukemia. N Engl J Med 1995; 333: pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478: 64–69. 1052–1057. 9 Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D et al. 2 Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50. 2011; 365: 1384–1395. 3 Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole- 10 Damm F, Nguyen-Khac F, Fontenay M, Bernard OA. Spliceosome and other novel genome sequencing identifies recurrent mutations in chronic lymphocytic leu- mutations in chronic lymphocytic leukemia and myeloid malignancies. Leukemia kaemia. Nature 2011; 475: 101–105. 2012; 26: 2027–2031. 4 Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L et al. Exome 11 Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in RNP machine. Cell 2009; 136: 701–718. chronic lymphocytic leukemia. Nat Genet 2012; 44: 47–52. 12 David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways 5 Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and programs unhinged. Genes Dev 2010; 24: 2343–2364. -

Transcriptional Regulation by Extracellular Signals 209
Cell, Vol. 80, 199-211, January 27, 1995, Copyright © 1995 by Cell Press Transcriptional Regulation Review by Extracellular Signals: Mechanisms and Specificity Caroline S. Hill and Richard Treisman Nuclear Translocation Transcription Laboratory In principle, regulated nuclear localization of transcription Imperial Cancer Research Fund factors can involve regulated activity of either nuclear lo- Lincoln's Inn Fields calization signals (NLSs) or cytoplasmic retention signals, London WC2A 3PX although no well-characterized case of the latter has yet England been reported. N LS activity, which is generally dependent on short regions of basic amino acids, can be regulated either by masking mechanisms or by phosphorylations Changes in cell behavior induced by extracellular signal- within the NLS itself (Hunter and Karin, 1992). For exam- ing molecules such as growth factors and cytokines re- ple, association with an inhibitory subunit masks the NLS quire execution of a complex program of transcriptional of NF-KB and its relatives (Figure 1; for review see Beg events. While the route followed by the intracellular signal and Baldwin, 1993), while an intramolecular mechanism from the cell membrane to its transcription factor targets may mask NLS activity in the heat shock regulatory factor can be traced in an increasing number of cases, how the HSF2 (Sheldon and Kingston, 1993). When transcription specificity of the transcriptional response of the cell to factor localization is dependent on regulated NLS activity, different stimuli is determined is much less clear. How- linkage to a constitutively acting NLS may be sufficient to ever, it is possible to understand at least in principle how render nuclear localization independent of signaling (Beg different stimuli can activate the same signal pathway yet et al., 1992). -

DNA·RNA Triple Helix Formation Can Function As a Cis-Acting Regulatory
DNA·RNA triple helix formation can function as a cis-acting regulatory mechanism at the human β-globin locus Zhuo Zhoua, Keith E. Gilesa,b,c, and Gary Felsenfelda,1 aLaboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892; bUniversity of Alabama at Birmingham Stem Cell Institute, University of Alabama at Birmingham, Birmingham, AL 35294; and cDepartment of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, Birmingham, AL 35294 Contributed by Gary Felsenfeld, February 4, 2019 (sent for review January 4, 2019; reviewed by James Douglas Engel and Sergei M. Mirkin) We have identified regulatory mechanisms in which an RNA tran- of the criteria necessary to establish the presence of a triplex script forms a DNA duplex·RNA triple helix with a gene or one of its structure, we first describe and characterize triplex formation at regulatory elements, suggesting potential auto-regulatory mecha- the FAU gene in human erythroid K562 cells. FAU encodes a nisms in vivo. We describe an interaction at the human β-globin protein that is a fusion containing fubi, a ubiquitin-like protein, locus, in which an RNA segment embedded in the second intron of and ribosomal protein S30. Although fubi function is unknown, the β-globin gene forms a DNA·RNA triplex with the HS2 sequence posttranslational processing produces S30, a component of the within the β-globin locus control region, a major regulator of glo- 40S ribosome. We used this system to refine methods necessary bin expression. We show in human K562 cells that the triplex is to detect triplex formation and to distinguish it from R-loop stable in vivo. -

NF-Κb Modifies the Mammalian Circadian Clock Through Interaction with the Core Clock Protein BMAL1
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. NF-κB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1 Yang Shen1, Wei Wang2, Mehari Endale1, Lauren J. Francey3, Rachel L. Harold4, David W. Hammers5, Zhiguang Huo6, Carrie L. Partch4, John B. Hogenesch3, Zhao-Hui Wu2, and Andrew C. Liu1* 1Department of Physiology and Functional Genomics, University of Florida College of Medicine; 2Department of Radiation Oncology and Center for Cancer Research, University of Tennessee Health Science Center; 3Divisions of Human Genetics and Immunobiology, Cincinnati Children's Hospital Medical Center; 4Department of Chemistry and Biochemistry, University of California Santa Cruz; 5Department of Pharmacology and Therapeutics, University of Florida College of Medicine; 6Department of Biostatistics, College of Public Health & Health Professions, University of Florida College of Medicine Running title: NF-κB modifies circadian clock * Correspondence: Andrew C. Liu, [email protected] Keywords: Circadian clock, inflammation, NF-κB, suprachiasmatic nucleus (SCN), BMAL1 Abbreviations: NF-κB, nuclear factor kappa B; SCN, suprachiasmatic nucleus; IKK, IκB kinase; IκBα, inhibitor of NF-κB; Luc, luciferase; RHD, Rel homology domain; CRD, C-terminal oscillation regulatory domain, TAD, transactivation domain 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

Transcritional Regulatory Networks Downstream of TAL1/SCL in T-Cell Acute Lymphoblastic Leukemia
Supplemental Materials Transcritional Regulatory Networks Downstream of TAL1/SCL in T-cell Acute Lymphoblastic Leukemia Palomero et al. Figure S1. Downregulation of TAL1 by RNA interference does not affect apoptosis in Jurkat cells. Figure S2. Scatter plots of Chromatin immunoprecipitations performed with TAL1#370 antibody in Jurkat cells. Figure S3. Target validation by gene-specific quantitative RT-PCR (qRT-PCR) Table S1. Genes regulated by TAL1 knockdown are identified as direct targets of this transcription factor using ChIP on chip. Table S2. Analysis of TAL1, E2A and HEB binding to TAL1 direct targets identified by ChIP on chip. Table S3. Previously described TAL1 targets. Figure S1. Downregulation of TAL1 by RNA interference does not affect apoptosis in Jurkat cells. Annexin V staining was used to quantify apoptosis rates in Jurkat cell clones expressing TAL1 shRNA (right panels) or a control shRNA (left panels). The percentage of apoptotic cells is indicated in the bottom right corner of each graph, while the dead cell percentage is indicated in the top right corner. PI: propidium iodide. Name Description p-value IFRD1 interferon-related developmental regulator 1 0.009927703 PCK2 phosphoenolpyruvate carboxykinase 2 (mitochondrial) 0.00445631 ATRX alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) 0.010847575 CDK6 cyclin-dependent kinase 6 0.044093956 Table S1. Genes regulated by TAL1 knockdown are identified as direct target of this transcription factor using ChIP on chip. The p-value determined by the error model applied to the ChIP on chip fluorescence data is indicated in the right column. Name Description Name Description E2A E2A HEB HEB TAL1 TAL1 Signal transduction-Receptor Transporters-lipids/small molecules MUC16 mucin 16 ABCC12 ATP-binding cassette, sub-fam. -

Function of Bromodomain and Extra-Terminal Motif Proteins (Bets) in Gata1-Mediated Transcription
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2015 Function of Bromodomain and Extra-Terminal Motif Proteins (bets) in Gata1-Mediated Transcription Aaron James Stonestrom University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Molecular Biology Commons, and the Pharmacology Commons Recommended Citation Stonestrom, Aaron James, "Function of Bromodomain and Extra-Terminal Motif Proteins (bets) in Gata1-Mediated Transcription" (2015). Publicly Accessible Penn Dissertations. 1148. https://repository.upenn.edu/edissertations/1148 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1148 For more information, please contact [email protected]. Function of Bromodomain and Extra-Terminal Motif Proteins (bets) in Gata1-Mediated Transcription Abstract Bromodomain and Extra-Terminal motif proteins (BETs) associate with acetylated histones and transcription factors. While pharmacologic inhibition of this ubiquitous protein family is an emerging therapeutic approach for neoplastic and inflammatory disease, the mechanisms through which BETs act remain largely uncharacterized. Here we explore the role of BETs in the physiologically relevant context of erythropoiesis driven by the transcription factor GATA1. First, we characterize functions of the BET family as a whole using a pharmacologic approach. We find that BETs are broadly required for GATA1-mediated transcriptional activation, but that repression is largely BET-independent. BETs support activation by facilitating both GATA1 occupancy and transcription downstream of its binding. Second, we test the specific olesr of BETs BRD2, BRD3, and BRD4 in GATA1-activated transcription. BRD2 and BRD4 are required for efficient anscriptionaltr activation by GATA1. Despite co-localizing with the great majority of GATA1 binding sites, we find that BRD3 is not equirr ed for GATA1-mediated transcriptional activation. -

ARID5B As a Critical Downstream Target of the TAL1 Complex That Activates the Oncogenic Transcriptional Program and Promotes T-Cell Leukemogenesis
Downloaded from genesdev.cshlp.org on October 10, 2021 - Published by Cold Spring Harbor Laboratory Press ARID5B as a critical downstream target of the TAL1 complex that activates the oncogenic transcriptional program and promotes T-cell leukemogenesis Wei Zhong Leong,1 Shi Hao Tan,1 Phuong Cao Thi Ngoc,1 Stella Amanda,1 Alice Wei Yee Yam,1 Wei-Siang Liau,1 Zhiyuan Gong,2 Lee N. Lawton,1 Daniel G. Tenen,1,3,4 and Takaomi Sanda1,4 1Cancer Science Institute of Singapore, National University of Singapore, 117599 Singapore; 2Department of Biological Sciences, National University of Singapore, 117543 Singapore; 3Harvard Medical School, Boston, Massachusetts 02215, USA; 4Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, 117599 Singapore The oncogenic transcription factor TAL1/SCL induces an aberrant transcriptional program in T-cell acute lym- phoblastic leukemia (T-ALL) cells. However, the critical factors that are directly activated by TAL1 and contribute to T-ALL pathogenesis are largely unknown. Here, we identified AT-rich interactive domain 5B (ARID5B) as a col- laborating oncogenic factor involved in the transcriptional program in T-ALL. ARID5B expression is down-regulated at the double-negative 2–4 stages in normal thymocytes, while it is induced by the TAL1 complex in human T-ALL cells. The enhancer located 135 kb upstream of the ARID5B gene locus is activated under a superenhancer in T-ALL cells but not in normal T cells. Notably, ARID5B-bound regions are associated predominantly with active tran- scription. ARID5B and TAL1 frequently co-occupy target genes and coordinately control their expression. -

Prox1regulates the Subtype-Specific Development of Caudal Ganglionic
The Journal of Neuroscience, September 16, 2015 • 35(37):12869–12889 • 12869 Development/Plasticity/Repair Prox1 Regulates the Subtype-Specific Development of Caudal Ganglionic Eminence-Derived GABAergic Cortical Interneurons X Goichi Miyoshi,1 Allison Young,1 Timothy Petros,1 Theofanis Karayannis,1 Melissa McKenzie Chang,1 Alfonso Lavado,2 Tomohiko Iwano,3 Miho Nakajima,4 Hiroki Taniguchi,5 Z. Josh Huang,5 XNathaniel Heintz,4 Guillermo Oliver,2 Fumio Matsuzaki,3 Robert P. Machold,1 and Gord Fishell1 1Department of Neuroscience and Physiology, NYU Neuroscience Institute, Smilow Research Center, New York University School of Medicine, New York, New York 10016, 2Department of Genetics & Tumor Cell Biology, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105, 3Laboratory for Cell Asymmetry, RIKEN Center for Developmental Biology, Kobe 650-0047, Japan, 4Laboratory of Molecular Biology, Howard Hughes Medical Institute, GENSAT Project, The Rockefeller University, New York, New York 10065, and 5Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724 Neurogliaform (RELNϩ) and bipolar (VIPϩ) GABAergic interneurons of the mammalian cerebral cortex provide critical inhibition locally within the superficial layers. While these subtypes are known to originate from the embryonic caudal ganglionic eminence (CGE), the specific genetic programs that direct their positioning, maturation, and integration into the cortical network have not been eluci- dated. Here, we report that in mice expression of the transcription factor Prox1 is selectively maintained in postmitotic CGE-derived cortical interneuron precursors and that loss of Prox1 impairs the integration of these cells into superficial layers. Moreover, Prox1 differentially regulates the postnatal maturation of each specific subtype originating from the CGE (RELN, Calb2/VIP, and VIP). -
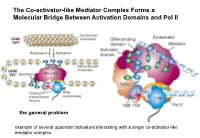
The Co-Activator-Like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II
The Co-activator-like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II Co-activator the general problem example of several upstream activators interacting with a single co-activator-like mediator complex Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P. Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION •Protein synthesized •Protein phosphorylated •Ligand binding •Release inhibitor •Change partner, etc RG Clerc April 6. 2016 Transcription factors as final effectors of the cellular signaling cascade Regulatory Mechanisms of Transcription Factor Function Genes X. Lewin B. editor Regulatory Mechanisms of Transcription Factor Function CREB FOXO NFAT Genes X. Lewin B. editor Body plan is constructed through interactions of the developmentally regulated homeotic gene expression anterior early expression posterior late expression high RA response low RA response hindbrain trunk Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Hoxb transcription factors mRNA distribution along the AP axis A staggered expression of the anterior border within somites is a property of the physical ordering along the chromosome: the colinearity Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Transcription control of the Hox genes: insight into colinear activation P A p p p p a p a p a a a a Tarchini B and Duboule D. Dev.Cell 10_93 (2006) correlation between linear arrangement along the chromosome and spatial -

E Proteins and ID Proteins: Helix-Loop-Helix Partners in Development and Disease
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Developmental Cell Review E Proteins and ID Proteins: Helix-Loop-Helix Partners in Development and Disease Lan-Hsin Wang1 and Nicholas E. Baker1,2,3,* 1Department of Genetics, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA 2Department of Developmental and Molecular Biology, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA 3Department of Ophthalmology and Visual Sciences, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA *Correspondence: [email protected] http://dx.doi.org/10.1016/j.devcel.2015.10.019 The basic Helix-Loop-Helix (bHLH) proteins represent a well-known class of transcriptional regulators. Many bHLH proteins act as heterodimers with members of a class of ubiquitous partners, the E proteins. A widely expressed class of inhibitory heterodimer partners—the Inhibitor of DNA-binding (ID) proteins—also exists. Genetic and molecular analyses in humans and in knockout mice implicate E proteins and ID proteins in a wide variety of diseases, belying the notion that they are non-specific partner proteins. Here, we explore relationships of E proteins and ID proteins to a variety of disease processes and highlight gaps in knowledge of disease mechanisms. E proteins and Inhibitor of DNA-binding (ID) proteins are widely conferring DNA-binding specificity and transcriptional activation expressed transcriptional regulators with very general functions. on heterodimers with the ubiquitous E proteins (Figure 1). They are implicated in diseases by evidence ranging from Another class of pervasive HLH proteins acts in opposition to confirmed Mendelian inheritance, association studies, and E proteins. -

The C-Rel Transcription Factor and B-Cell Proliferation: a Deal with the Devil
Oncogene (2004) 23, 2275–2286 & 2004 Nature Publishing Group All rights reserved 0950-9232/04 $25.00 www.nature.com/onc REVIEW The c-Rel transcription factor and B-cell proliferation: a deal with the devil Thomas D Gilmore*,1, Demetrios Kalaitzidis1, Mei-Chih Liang1 and Daniel T Starczynowski1 1Department of Biology, Boston University, 5 Cummington Street, Boston, MA 02215, USA Activation of the Rel/NF-jB signal transduction pathway called the Rel homology domain, which has sequences has been associated with a varietyof animal and human required for DNA binding, dimerization, nuclear malignancies. However, among the Rel/NF-jB family localization, and inhibitor (IkB) binding. Sequences members, onlyc-Rel has been consistentlyshown to be C-terminal to the Rel homology domain in c-Rel, RelA, able to malignantlytransform cells in culture. In addition, and RelB contain transcriptional activation domains, c-rel has been activated bya retroviral promoter insertion and therefore dimers that contain these Rel/NF-kB in an avian B-cell lymphoma, and amplifications of REL members usually increase transcription of target genes. (human c-rel) are frequentlyseen in Hodgkin’s lympho- Although the Rel homology domain sequences of c-Rel mas and diffuse large B-cell lymphomas, and in some proteins across species are highly conserved, their follicular and mediastinal B-cell lymphomas. Phenotypic C-terminal transactivation domains are not. For exam- analysis of c-rel knockout mice demonstrates that c-Rel ple, the Rel homology domains of chicken and human has a normal role in B-cell proliferation and survival; c-Rel are approximately 85% identical, whereas their moreover, c-Rel nuclear activityis required for B-cell C-terminal transactivation domains are only about 10% development. -

The Role of Ubiquitination in NF-Κb Signaling During Virus Infection
viruses Review The Role of Ubiquitination in NF-κB Signaling during Virus Infection Kun Song and Shitao Li * Department of Microbiology and Immunology, Tulane University, New Orleans, LA 70112, USA; [email protected] * Correspondence: [email protected] Abstract: The nuclear factor κB (NF-κB) family are the master transcription factors that control cell proliferation, apoptosis, the expression of interferons and proinflammatory factors, and viral infection. During viral infection, host innate immune system senses viral products, such as viral nucleic acids, to activate innate defense pathways, including the NF-κB signaling axis, thereby inhibiting viral infection. In these NF-κB signaling pathways, diverse types of ubiquitination have been shown to participate in different steps of the signal cascades. Recent advances find that viruses also modulate the ubiquitination in NF-κB signaling pathways to activate viral gene expression or inhibit host NF-κB activation and inflammation, thereby facilitating viral infection. Understanding the role of ubiquitination in NF-κB signaling during viral infection will advance our knowledge of regulatory mechanisms of NF-κB signaling and pave the avenue for potential antiviral therapeutics. Thus, here we systematically review the ubiquitination in NF-κB signaling, delineate how viruses modulate the NF-κB signaling via ubiquitination and discuss the potential future directions. Keywords: NF-κB; polyubiquitination; linear ubiquitination; inflammation; host defense; viral infection Citation: Song, K.; Li, S. The Role of 1. Introduction Ubiquitination in NF-κB Signaling The nuclear factor κB (NF-κB) is a small family of five transcription factors, including during Virus Infection. Viruses 2021, RelA (also known as p65), RelB, c-Rel, p50 and p52 [1].