Transcriptional Regulation by Extracellular Signals 209
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New Editor on Journal of Cell Science Michael Way (Editor-In-Chief)
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2019) 132, jcs229740. doi:10.1242/jcs.229740 EDITORIAL New Editor on Journal of Cell Science Michael Way (Editor-in-Chief) As someone who has worked on things related to the actin cytoskeleton my whole research career, the nucleus was not something I paid much attention to. Yes, there were scattered historical reports of actin in the nucleus long before I started my PhD, but no one believed actin was really there of course – it was all an artefact of fixation, you know. Nuclear actin was taboo and no one talked about it at the meetings I went to as a student and postdoc. How wrong we were – today nuclear actin is alive and kicking, although there are definitely more questions than answers concerning what it is actually doing there. We now appreciate that the nucleus contains a wide assortment of proteins associated with the cytoplasmic actin cytoskeleton including myosin motors and actin nucleators such as the Arp2/3 complex. In addition, it should not be forgotten that many chromatin-associated complexes including SWI/SNF and INO80/ SWR also contain multiple actin-related proteins, as well as actin itself. It strikes me that maybe we should all be paying more attention to the nucleus and not just because it contains my favourite proteins! Maybe that’s why, in recent years, we’ve been seeing more submissions to JCS that are focused on different aspects of the nucleus and that traditionally appeared in journals with ‘molecular’ in their titles. -

NF-Κb Modifies the Mammalian Circadian Clock Through Interaction with the Core Clock Protein BMAL1
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. NF-κB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1 Yang Shen1, Wei Wang2, Mehari Endale1, Lauren J. Francey3, Rachel L. Harold4, David W. Hammers5, Zhiguang Huo6, Carrie L. Partch4, John B. Hogenesch3, Zhao-Hui Wu2, and Andrew C. Liu1* 1Department of Physiology and Functional Genomics, University of Florida College of Medicine; 2Department of Radiation Oncology and Center for Cancer Research, University of Tennessee Health Science Center; 3Divisions of Human Genetics and Immunobiology, Cincinnati Children's Hospital Medical Center; 4Department of Chemistry and Biochemistry, University of California Santa Cruz; 5Department of Pharmacology and Therapeutics, University of Florida College of Medicine; 6Department of Biostatistics, College of Public Health & Health Professions, University of Florida College of Medicine Running title: NF-κB modifies circadian clock * Correspondence: Andrew C. Liu, [email protected] Keywords: Circadian clock, inflammation, NF-κB, suprachiasmatic nucleus (SCN), BMAL1 Abbreviations: NF-κB, nuclear factor kappa B; SCN, suprachiasmatic nucleus; IKK, IκB kinase; IκBα, inhibitor of NF-κB; Luc, luciferase; RHD, Rel homology domain; CRD, C-terminal oscillation regulatory domain, TAD, transactivation domain 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

Homeobox Gene Expression Profile in Human Hematopoietic Multipotent
Leukemia (2003) 17, 1157–1163 & 2003 Nature Publishing Group All rights reserved 0887-6924/03 $25.00 www.nature.com/leu Homeobox gene expression profile in human hematopoietic multipotent stem cells and T-cell progenitors: implications for human T-cell development T Taghon1, K Thys1, M De Smedt1, F Weerkamp2, FJT Staal2, J Plum1 and G Leclercq1 1Department of Clinical Chemistry, Microbiology and Immunology, Ghent University Hospital, Ghent, Belgium; and 2Department of Immunology, Erasmus Medical Center, Rotterdam, The Netherlands Class I homeobox (HOX) genes comprise a large family of implicated in this transformation proces.14 The HOX-C locus transcription factors that have been implicated in normal and has been primarily implicated in lymphomas.15 malignant hematopoiesis. However, data on their expression or function during T-cell development is limited. Using degener- Hematopoietic cells are derived from stem cells that reside in ated RT-PCR and Affymetrix microarray analysis, we analyzed fetal liver (FL) in the embryo and in the adult bone marrow the expression pattern of this gene family in human multipotent (ABM), which have the unique ability to self-renew and thereby stem cells from fetal liver (FL) and adult bone marrow (ABM), provide a life-long supply of blood cells. T lymphocytes are a and in T-cell progenitors from child thymus. We show that FL specific type of hematopoietic cells that play a major role in the and ABM stem cells are similar in terms of HOX gene immune system. They develop through a well-defined order of expression, but significant differences were observed between differentiation steps in the thymus.16 Several transcription these two cell types and child thymocytes. -
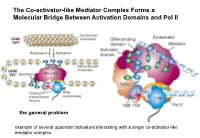
The Co-Activator-Like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II
The Co-activator-like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II Co-activator the general problem example of several upstream activators interacting with a single co-activator-like mediator complex Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P. Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION •Protein synthesized •Protein phosphorylated •Ligand binding •Release inhibitor •Change partner, etc RG Clerc April 6. 2016 Transcription factors as final effectors of the cellular signaling cascade Regulatory Mechanisms of Transcription Factor Function Genes X. Lewin B. editor Regulatory Mechanisms of Transcription Factor Function CREB FOXO NFAT Genes X. Lewin B. editor Body plan is constructed through interactions of the developmentally regulated homeotic gene expression anterior early expression posterior late expression high RA response low RA response hindbrain trunk Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Hoxb transcription factors mRNA distribution along the AP axis A staggered expression of the anterior border within somites is a property of the physical ordering along the chromosome: the colinearity Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Transcription control of the Hox genes: insight into colinear activation P A p p p p a p a p a a a a Tarchini B and Duboule D. Dev.Cell 10_93 (2006) correlation between linear arrangement along the chromosome and spatial -

The C-Rel Transcription Factor and B-Cell Proliferation: a Deal with the Devil
Oncogene (2004) 23, 2275–2286 & 2004 Nature Publishing Group All rights reserved 0950-9232/04 $25.00 www.nature.com/onc REVIEW The c-Rel transcription factor and B-cell proliferation: a deal with the devil Thomas D Gilmore*,1, Demetrios Kalaitzidis1, Mei-Chih Liang1 and Daniel T Starczynowski1 1Department of Biology, Boston University, 5 Cummington Street, Boston, MA 02215, USA Activation of the Rel/NF-jB signal transduction pathway called the Rel homology domain, which has sequences has been associated with a varietyof animal and human required for DNA binding, dimerization, nuclear malignancies. However, among the Rel/NF-jB family localization, and inhibitor (IkB) binding. Sequences members, onlyc-Rel has been consistentlyshown to be C-terminal to the Rel homology domain in c-Rel, RelA, able to malignantlytransform cells in culture. In addition, and RelB contain transcriptional activation domains, c-rel has been activated bya retroviral promoter insertion and therefore dimers that contain these Rel/NF-kB in an avian B-cell lymphoma, and amplifications of REL members usually increase transcription of target genes. (human c-rel) are frequentlyseen in Hodgkin’s lympho- Although the Rel homology domain sequences of c-Rel mas and diffuse large B-cell lymphomas, and in some proteins across species are highly conserved, their follicular and mediastinal B-cell lymphomas. Phenotypic C-terminal transactivation domains are not. For exam- analysis of c-rel knockout mice demonstrates that c-Rel ple, the Rel homology domains of chicken and human has a normal role in B-cell proliferation and survival; c-Rel are approximately 85% identical, whereas their moreover, c-Rel nuclear activityis required for B-cell C-terminal transactivation domains are only about 10% development. -

The Role of Ubiquitination in NF-Κb Signaling During Virus Infection
viruses Review The Role of Ubiquitination in NF-κB Signaling during Virus Infection Kun Song and Shitao Li * Department of Microbiology and Immunology, Tulane University, New Orleans, LA 70112, USA; [email protected] * Correspondence: [email protected] Abstract: The nuclear factor κB (NF-κB) family are the master transcription factors that control cell proliferation, apoptosis, the expression of interferons and proinflammatory factors, and viral infection. During viral infection, host innate immune system senses viral products, such as viral nucleic acids, to activate innate defense pathways, including the NF-κB signaling axis, thereby inhibiting viral infection. In these NF-κB signaling pathways, diverse types of ubiquitination have been shown to participate in different steps of the signal cascades. Recent advances find that viruses also modulate the ubiquitination in NF-κB signaling pathways to activate viral gene expression or inhibit host NF-κB activation and inflammation, thereby facilitating viral infection. Understanding the role of ubiquitination in NF-κB signaling during viral infection will advance our knowledge of regulatory mechanisms of NF-κB signaling and pave the avenue for potential antiviral therapeutics. Thus, here we systematically review the ubiquitination in NF-κB signaling, delineate how viruses modulate the NF-κB signaling via ubiquitination and discuss the potential future directions. Keywords: NF-κB; polyubiquitination; linear ubiquitination; inflammation; host defense; viral infection Citation: Song, K.; Li, S. The Role of 1. Introduction Ubiquitination in NF-κB Signaling The nuclear factor κB (NF-κB) is a small family of five transcription factors, including during Virus Infection. Viruses 2021, RelA (also known as p65), RelB, c-Rel, p50 and p52 [1]. -

The Role of WH2-Containing Proteins in Regulating Actin-MRTF-SRF-Mediated Transcription
The Role of WH2-containing Proteins in Regulating Actin-MRTF-SRF-mediated Transcription Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) der Naturwissenschaftlichen Fakultät I – Biowissenschaften - der Martin-Luther-Universität Halle-Wittenberg vorgelegt von Frau Julia Weißbach geboren am 24.06.1987 in Querfurt Gutachter: Prof. Guido Posern Prof. Mechthild Hatzfeld Prof. Bernd Knöll Verteidigungsdatum: 15. November 2017 Content I Summary ...................................................................................................................... 3 Zusammenfassung ....................................................................................................... 4 II Introduction ............................................................................................................... 5 II.1 Serum Response Factor - SRF ............................................................................... 5 II.2 Myocardin-related Transcription Factors – MRTF-A/-B ...................................... 6 II.3 Actin and Actin-Binding Proteins .......................................................................... 9 II.4 Nucleation Promoting Factors – NPF .................................................................. 12 II.4.1 Neuronal Wiskott-Aldrich Syndrome Protein – N-WASP ........................... 14 II.4.2 WASP Family Verprolin-homologous Protein 2 - WAVE2 ......................... 15 II.4.3 Junction-mediating and Regulatory Protein - JMY ...................................... 15 -

Pax6 During Visual System Development
Hedgehog-dependent E3-ligase Midline1 regulates ubiquitin-mediated proteasomal degradation of Pax6 during visual system development Thorsten Pfirrmanna,1, Enrico Jandta,1, Swantje Ranfta,b, Ashwin Lokapallya, Herbert Neuhausa, Muriel Perronc, and Thomas Hollemanna,2 aInstitute for Physiological Chemistry, University of Halle-Wittenberg, 06114 Halle, Germany; bGynecological Hospital, University Medical Center Mannheim, 68167 Mannheim, Germany; and cParis-Saclay Institute of Neuroscience, CNRS, Univ Paris Sud, Université Paris-Saclay, 91405 Orsay, France Edited by Richard M. Harland, University of California, Berkeley, CA, and approved July 19, 2016 (received for review January 16, 2016) Pax6 is a key transcription factor involved in eye, brain, and pancreas remains unclear how Pax6 protein is removed from the eyestalk development. Although pax6 is expressed in the whole prospective territory on time. Some authors report the regulation of Pax6 retinal field, subsequently its expression becomes restricted to the activity by posttranslational modifications (21–23), and most optic cup by reciprocal transcriptional repression of pax6 and pax2. interestingly, Tuoc et al. showed that in cortical progenitor cells, However, it remains unclear how Pax6 protein is removed from the Pax6 protein is degraded by the proteasome mediated by Trim11 eyestalk territory on time. Here, we report that Mid1, a member of (24). However, the existence of similar mechanisms leading to the RBCC/TRIM E3 ligase family, which was first identified in patients the development of the visual system is not known. with the X-chromosome–linked Opitz BBB/G (OS) syndrome, inter- The data of our present study show that Midline1 (Mid1) acts with Pax6. We found that the forming eyestalk is a major do- serves as one of these links. -

And C-Terminal Domains of Relb Are Required for Full Transactivation
MOLECULAR AND CELLULAR BIOLOGY, Mar. 1993, p. 1572-1582 Vol. 13, No. 3 0270-7306/93/031572-11$02.00/0 Copyright © 1993, American Society for Microbiology Both N- and C-Terminal Domains of RelB Are Required for Full Transactivation: Role of the N-Terminal Leucine Zipper-Like Motif PAWEL DOBRZANSKI, ROLF-PETER RYSECK, AND RODRIGO BRAVO* Department ofMolecular Biology, Bristol-Myers Squibb Pharmaceutical Research Institute, P.O. Box 4000, Princeton, New Jersey 08543-4000 Received 18 September 1992/Returned for modification 11 November 1992/Accepted 12 December 1992 RelB, a member of the Rel family of transcription factors, can stimulate promoter activity in the presence of p50-NF-KB or p5OB/p49-NF-KB in mammalian cells. Transcriptional activation analysis reveals that the N and C termini of RelB are required for full transactivation in the presence of p50-NF-KB. RelB/p50-NF-KB hybrid molecules containing the Rel homology domain of p50-NF-KB and the N and C termini of RelB have high transcriptional activity compared with wild-type p50-NF-KB. The N and C termini of RelB cooperate in transactivation in cis or trans configuration. Alterations in the structure of the leucine zipper-like motif present in the N terminus of RelB significantly decrease the transcriptional capacity of RelB and of different RelB/p50-NF-KB hybrid molecules. Serum stimulation of quiescent fibroblasts induces the regulated by the inhibitory factor cactus (28). The different expression of more than 100 genes, including several proto- homodimers and heterodimers formed between the members oncogenes encoding for transcriptional factors belonging to of the Rel family regulate gene expression in vivo (20, 22, 42) the Jun, Fos, and Rel families (2, 11, 13, 24, 34-36, 38). -

TGF-B Uses a Novel Mode of Receptor Activation to Phosphorylate SMAD1
RESEARCH ARTICLE TGF-b uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition Anassuya Ramachandran1, Pedro Viza´ n1†, Debipriya Das1‡, Probir Chakravarty2, Janis Vogt3, Katherine W Rogers4, Patrick Mu¨ ller4, Andrew P Hinck5, Gopal P Sapkota3, Caroline S Hill1* 1Developmental Signalling Laboratory, The Francis Crick Institute, London, United Kingdom; 2Bioinformatics and Biostatistics Facility, The Francis Crick Institute, London, United Kingdom; 3Medical Research Council Protein Phosphorylation and Ubiquitylation Unit, University of Dundee, Dundee, United Kingdom; 4Friedrich Miescher Laboratory of the Max Planck Society, Tu¨ bingen, Germany; 5Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, United States Abstract The best characterized signaling pathway downstream of transforming growth factor b (TGF-b) is through SMAD2 and SMAD3. However, TGF-b also induces phosphorylation of SMAD1 *For correspondence: and SMAD5, but the mechanism of this phosphorylation and its functional relevance is not known. [email protected] Here, we show that TGF-b-induced SMAD1/5 phosphorylation requires members of two classes of type I receptor, TGFBR1 and ACVR1, and establish a new paradigm for receptor activation where Present address: †Center for TGFBR1 phosphorylates and activates ACVR1, which phosphorylates SMAD1/5. We demonstrate Genomic Regulation, Barcelona, ‡ the biological significance of this pathway by showing that approximately a quarter of the TGF-b- Spain; Flow Cytometry, The Francis Crick Institute, London, induced transcriptome depends on SMAD1/5 signaling, with major early transcriptional targets ID United Kingdom being the genes. Finally, we show that TGF-b-induced epithelial-to-mesenchymal transition requires signaling via both the SMAD3 and SMAD1/5 pathways, with SMAD1/5 signaling being Competing interests: The essential to induce ID1. -

AP-1 in Cell Proliferation and Survival
Oncogene (2001) 20, 2390 ± 2400 ã 2001 Nature Publishing Group All rights reserved 0950 ± 9232/01 $15.00 www.nature.com/onc AP-1 in cell proliferation and survival Eitan Shaulian1 and Michael Karin*,1 1Laboratory of Gene Regulation and Signal Transduction, Department of Pharmacology, University of California San Diego, 9500 Gilman Drive, La Jolla, California, CA 92093-0636, USA A plethora of physiological and pathological stimuli extensively discussed previously (Angel and Karin, induce and activate a group of DNA binding proteins 1991; Karin, 1995). that form AP-1 dimers. These proteins include the Jun, The mammalian AP-1 proteins are homodimers and Fos and ATF subgroups of transcription factors. Recent heterodimers composed of basic region-leucine zipper studies using cells and mice de®cient in individual AP-1 (bZIP) proteins that belong to the Jun (c-Jun, JunB proteins have begun to shed light on their physiological and JunD), Fos (c-Fos, FosB, Fra-1 and Fra-2), Jun functions in the control of cell proliferation, neoplastic dimerization partners (JDP1 and JDP2) and the closely transformation and apoptosis. Above all such studies related activating transcription factors (ATF2, LRF1/ have identi®ed some of the target genes that mediate the ATF3 and B-ATF) subfamilies (reviewed by (Angel eects of AP-1 proteins on cell proliferation and death. and Karin, 1991; Aronheim et al., 1997; Karin et al., There is evidence that AP-1 proteins, mostly those that 1997; Liebermann et al., 1998; Wisdom, 1999). In belong to the Jun group, control cell life and death addition, some of the Maf proteins (v-Maf, c-Maf and through their ability to regulate the expression and Nrl) can heterodimerize with c-Jun or c-Fos (Nishiza- function of cell cycle regulators such as Cyclin D1, p53, wa et al., 1989; Swaroop et al., 1992), whereas other p21cip1/waf1, p19ARF and p16. -

Direct Interaction Between Estrogen Receptor and NF-B in the Nucleus Of
Direct interaction between estrogen receptor α and NF-κB in the nucleus of living cells Monique E. Quaedackers, Christina E. van den Brink, Paul T. van der Saag, Leon G.J. Tertoolen To cite this version: Monique E. Quaedackers, Christina E. van den Brink, Paul T. van der Saag, Leon G.J. Tertoolen. Direct interaction between estrogen receptor α and NF-κB in the nucleus of living cells. Molecular and Cellular Endocrinology, Elsevier, 2007, 273 (1-2), pp.42. 10.1016/j.mce.2007.05.002. hal-00531925 HAL Id: hal-00531925 https://hal.archives-ouvertes.fr/hal-00531925 Submitted on 4 Nov 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Accepted Manuscript Title: Direct interaction between estrogen receptor ␣ and NF-B in the nucleus of living cells Authors: Monique E. Quaedackers, Christina E. van den Brink, Paul T. van der Saag, Leon G.J. Tertoolen PII: S0303-7207(07)00185-2 DOI: doi:10.1016/j.mce.2007.05.002 Reference: MCE 6650 To appear in: Molecular and Cellular Endocrinology Received date: 27-3-2007 Revised date: 7-5-2007 Accepted date: 8-5-2007 Please cite this article as: Quaedackers, M.E., van den Brink, C.E., van der Saag, P.T., Tertoolen, L.G.J., Direct interaction between estrogen receptor ␣ and NF- B in the nucleus of living cells, Molecular and Cellular Endocrinology (2007), doi:10.1016/j.mce.2007.05.002 This is a PDF file of an unedited manuscript that has been accepted for publication.