Insect Transcription Factors: a Landscape of Their Structures and Biological Functions in Drosophila and Beyond
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Downloading Them Directly from the GO
bioRxiv preprint doi: https://doi.org/10.1101/284828; this version posted March 26, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: Evolution of the D. melanogaster chromatin landscape and its associated proteins Authors and affiliations: Elise Parey(1, 2) and Anton Crombach*(1,3) (1) Center for Interdisciplinary Research in Biology (CIRB), Collè!e de France, C#RS, IN$ERM, P$& Uni(ersit) Paris, 75005 Paris, France (2) (c-rrent address) Institut de Biologie de l’Ecole Normale Supérieure (IBENS), Ecole Normale Supérieure, CNRS, INSERM, P$& Uni(ersit) Paris, 75005 Paris, France (3) (current address) Inria, Antenne Lyon La Doua, 69603 Villeurbanne, France Author for Correspondence (*): Anton Crombach, Center for Interdisciplinary Research in Biology (CIRB), Collè!e de France, CNRS, I#$ERM, P$& Uni(ersit) Paris, 75005 Paris, "rance, anton.crombach@colle!e0de-france.fr Keywords: phylogenomics, chromatin-associated proteins, chromatin types, histone modi1cations, centromere dri(e, D. melanogaster. 1 of 46 bioRxiv preprint doi: https://doi.org/10.1101/284828; this version posted March 26, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract (221w, max 250w) In the nucleus of eukaryotic cells, !enomic 3#A associates 4ith numerous protein comple5es and RNAs, forming the chromatin landscape. -

Mir-2 Family Regulates Insect Metamorphosis by Controlling the Juvenile Hormone Signaling Pathway
MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway Jesus Lozanoa, Raúl Montañeza,b, and Xavier Bellesa,1 aInstitut de Biologia Evolutiva, Consejo Superior de Investigaciones Científicas, Universitat Pompeu Fabra, 08003 Barcelona, Spain; and bInstitució Catalana de Recerca i Estudis Avançats–Complex Systems Laboratory, Universitat Pompeu Fabra, 08003 Barcelona, Spain Edited by Lynn M. Riddiford, Howard Hughes Medical Institute Janelia Farm Research Campus, Ashburn, VA, and approved February 4, 2015 (received for review September 25, 2014) In 2009 we reported that depletion of Dicer-1, the enzyme that we used the cockroach Blattella germanica as a model, because catalyzes the final step of miRNA biosynthesis, prevents meta- it displays the more primitive hemimetabolan mode of meta- morphosis in Blattella germanica. However, the precise regulatory morphosis, where the juvenile stages already have the adult body roles of miRNAs in the process have remained elusive. In the pres- plan, thus making the transition from nymph to adult much less ent work, we have observed that Dicer-1 depletion results in an dramatic than in holometabolan species. In both hemimetabolan increase of mRNA levels of Krüppel homolog 1 (Kr-h1), a juvenile and holometabolan modes the regulation of metamorphosis is ba- hormone-dependent transcription factor that represses metamor- sically ensured by two hormones, the molting hormone (generally phosis, and that depletion of Kr-h1 expression in Dicer-1 knock- 20-hydroxyecdysone), which triggers nonmetamorphic and meta- down individuals rescues metamorphosis. We have also found morphic molts, and the juvenile hormone (JH) that represses the that the 3′UTR of Kr-h1 mRNA contains a functional binding site metamorphic character of the molts (21). -

Transcriptional Regulation by Extracellular Signals 209
Cell, Vol. 80, 199-211, January 27, 1995, Copyright © 1995 by Cell Press Transcriptional Regulation Review by Extracellular Signals: Mechanisms and Specificity Caroline S. Hill and Richard Treisman Nuclear Translocation Transcription Laboratory In principle, regulated nuclear localization of transcription Imperial Cancer Research Fund factors can involve regulated activity of either nuclear lo- Lincoln's Inn Fields calization signals (NLSs) or cytoplasmic retention signals, London WC2A 3PX although no well-characterized case of the latter has yet England been reported. N LS activity, which is generally dependent on short regions of basic amino acids, can be regulated either by masking mechanisms or by phosphorylations Changes in cell behavior induced by extracellular signal- within the NLS itself (Hunter and Karin, 1992). For exam- ing molecules such as growth factors and cytokines re- ple, association with an inhibitory subunit masks the NLS quire execution of a complex program of transcriptional of NF-KB and its relatives (Figure 1; for review see Beg events. While the route followed by the intracellular signal and Baldwin, 1993), while an intramolecular mechanism from the cell membrane to its transcription factor targets may mask NLS activity in the heat shock regulatory factor can be traced in an increasing number of cases, how the HSF2 (Sheldon and Kingston, 1993). When transcription specificity of the transcriptional response of the cell to factor localization is dependent on regulated NLS activity, different stimuli is determined is much less clear. How- linkage to a constitutively acting NLS may be sufficient to ever, it is possible to understand at least in principle how render nuclear localization independent of signaling (Beg different stimuli can activate the same signal pathway yet et al., 1992). -

NF-Κb Modifies the Mammalian Circadian Clock Through Interaction with the Core Clock Protein BMAL1
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. NF-κB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1 Yang Shen1, Wei Wang2, Mehari Endale1, Lauren J. Francey3, Rachel L. Harold4, David W. Hammers5, Zhiguang Huo6, Carrie L. Partch4, John B. Hogenesch3, Zhao-Hui Wu2, and Andrew C. Liu1* 1Department of Physiology and Functional Genomics, University of Florida College of Medicine; 2Department of Radiation Oncology and Center for Cancer Research, University of Tennessee Health Science Center; 3Divisions of Human Genetics and Immunobiology, Cincinnati Children's Hospital Medical Center; 4Department of Chemistry and Biochemistry, University of California Santa Cruz; 5Department of Pharmacology and Therapeutics, University of Florida College of Medicine; 6Department of Biostatistics, College of Public Health & Health Professions, University of Florida College of Medicine Running title: NF-κB modifies circadian clock * Correspondence: Andrew C. Liu, [email protected] Keywords: Circadian clock, inflammation, NF-κB, suprachiasmatic nucleus (SCN), BMAL1 Abbreviations: NF-κB, nuclear factor kappa B; SCN, suprachiasmatic nucleus; IKK, IκB kinase; IκBα, inhibitor of NF-κB; Luc, luciferase; RHD, Rel homology domain; CRD, C-terminal oscillation regulatory domain, TAD, transactivation domain 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.06.285254; this version posted September 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. -

Climatic Variation of Supercooling Point in the Linden Bug Pyrrhocoris Apterus (Heteroptera: Pyrrhocoridae)
insects Article Climatic Variation of Supercooling Point in the Linden Bug Pyrrhocoris apterus (Heteroptera: Pyrrhocoridae) Tomáš Ditrich 1,* ,Václav Janda 1, Hana Vanˇeˇcková 2 and David Doležel 2 1 Faculty of Education, University of South Bohemia, Branisovska 31a, 37005 Ceske Budejovice, Czech Republic; [email protected] 2 Biology Center, Academy of Sciences of the Czech Republic, 37005 Ceske Budejovice, Czech Republic; [email protected] (H.V.); [email protected] (D.D.) * Correspondence: [email protected]; Tel.: +420-387-773-014 Received: 30 June 2018; Accepted: 15 October 2018; Published: 19 October 2018 Abstract: Cold tolerance is often one of the key components of insect fitness, but the association between climatic conditions and supercooling capacity is poorly understood. We tested 16 lines originating from geographically different populations of the linden bug Pyrrhocoris apterus for their cold tolerance, determined as the supercooling point (SCP). The supercooling point was generally well explained by the climatic conditions of the population’s origin, as the best predictor—winter minimum temperature—explained 85% of the average SCP variation between populations. The supercooling capacity of P. apterus is strongly correlated with climatic conditions, which support the usage of SCP as an appropriate metric of cold tolerance in this species. Keywords: cold tolerance; supercooling point; overwintering; diapause 1. Introduction Population dynamics of temperate insects can be crucially affected by their survival rates during overwintering [1]. Cold tolerance, a key determinant of insect survival during winter, is thus an important research topic for insect ecologists and physiologists. This importance has increased with the recent climate changes around the world, especially in the Northern Hemisphere [2,3]. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Evolution of the D. Melanogaster Chromatin Landscape and Its Associated Proteins
bioRxiv preprint doi: https://doi.org/10.1101/284828; this version posted January 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: Evolution of the D. melanogaster chromatin landscape and its associated proteins Authors and affiliations: Elise Parey(1, 2) and Anton Crombach*(1,3,4) (1) Center for Interdisciplinary Research in Biology (CIRB), Collège de France, CNRS, INSERM, PSL Université Paris, 75005 Paris, France (2) (current address) Institut de Biologie de l’Ecole Normale Supérieure (IBENS), Ecole Normale Supérieure, CNRS, INSERM, PSL Université Paris, 75005 Paris, France (3) (current address) Inria, Antenne Lyon La Doua, 69603 Villeurbanne, France (4) Université de Lyon, INSA-Lyon, LIRIS, UMR 5205, 69621 Villeurbanne, France Author for Correspondence (*): Anton Crombach, Inria, Antenne Lyon La Doua, 69603 Villeurbanne, France, [email protected] 1 of 49 bioRxiv preprint doi: https://doi.org/10.1101/284828; this version posted January 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract (240w, max 250w) In the nucleus of eukaryotic cells, genomic DNA associates with numerous protein complexes and RNAs, forming the chromatin landscape. Through a genome-wide study of chromatin- associated proteins in Drosophila cells, five major chromatin types were identified as a refinement of the traditional binary division into hetero- and euchromatin. -
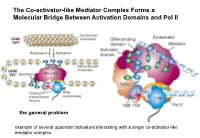
The Co-Activator-Like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II
The Co-activator-like Mediator Complex Forms a Molecular Bridge Between Activation Domains and Pol II Co-activator the general problem example of several upstream activators interacting with a single co-activator-like mediator complex Transcription Regulation And Gene Expression in Eukaryotes FS 2016 Graduate Course G2 P. Matthias and RG Clerc Pharmazentrum Hörsaal 2 16h15-18h00 REGULATORY MECHANISMS OF TRANSCRIPTION FACTOR FUNCTION •Protein synthesized •Protein phosphorylated •Ligand binding •Release inhibitor •Change partner, etc RG Clerc April 6. 2016 Transcription factors as final effectors of the cellular signaling cascade Regulatory Mechanisms of Transcription Factor Function Genes X. Lewin B. editor Regulatory Mechanisms of Transcription Factor Function CREB FOXO NFAT Genes X. Lewin B. editor Body plan is constructed through interactions of the developmentally regulated homeotic gene expression anterior early expression posterior late expression high RA response low RA response hindbrain trunk Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Hoxb transcription factors mRNA distribution along the AP axis A staggered expression of the anterior border within somites is a property of the physical ordering along the chromosome: the colinearity Alexander T and Krumlauf R. Ann.Rev.Cell.Dev.Biol: 25_431 (2009) Transcription control of the Hox genes: insight into colinear activation P A p p p p a p a p a a a a Tarchini B and Duboule D. Dev.Cell 10_93 (2006) correlation between linear arrangement along the chromosome and spatial -

Pyrrhocoris Apterus (Linnaeus, 1758) - a New Record of Firebug (Hemiptera, Pyrrhocoridae) from Malta
Correspondence BULL. ENT. SOC. MALTA (2019) Vol. 10 : 106–107 DOI: 10.17387/BULLENTSOCMALTA.2019.12 Pyrrhocoris apterus (Linnaeus, 1758) - a new record of firebug (Hemiptera, Pyrrhocoridae) from Malta Thomas CASSAR1 True bugs of the family Pyrrhocoridae, more commonly known as red bugs, cotton stainers or (in the case of Pyrrhocoris apterus) firebugs, are represented by 43 species from 13 genera in the Palaearctic (AUKEMA & RIEGER, 2001). The Heteroptera of the Maltese Islands have been relatively well-studied, with 141 species recorded (CUESTA SEGURA et al., 2010; CARAPEZZA & MIFSUD, 2015; 2016). Of these, only one species belongs to the family Pyrrhocoridae - Scantius aegyptius aegyptius. However, specimens collected in summer of 2018 were confirmed to bePyrrhocoris apterus, a new Pyrrhocorid record for Malta. Pyrrhocoris apterus (Linnaeus, 1758) Material examined: Malta, Ħaż-Żebbuġ, 20.vi.2018, leg. T. Cassar (1 macropterous); Rabat, Chadwick Lakes, 16.ix.2018, leg. T. Cassar (1 brachypterous); Rabat, Chadwick Lakes, 19.ix.2018, leg. T. Cassar (1 brachypterous). Notes: Pyrrhocoris apterus has a predominantly Palaearctic distribution, being present from the Iberian Peninsula eastwards into Siberia and China, including most of central and southern Europe. In controlled conditions, eggs take seven and a half days to hatch (MATOLÍN, 1973). P. apterus goes through five larval instars, the last of which lasts the longest – typically about seven days R( IZKI & SLÁMA, 1968). Adults can be either brachypterous or macropterous, though various intermediate morphs exist. Brachypters’ wings are reduced to vestigial scales whilst macropterous individuals have well-developed pairs of wings which extend to the abdomen tip or past it (SEIDENSTÜCKER, 1953). -

IISER Pune Annual Report 2015-16 Chairperson Pune, India Prof
dm{f©H$ à{VdoXZ Annual Report 2015-16 ¼ããäÌãÓ¾ã ãä¶ã¹ã¥ã †Ìãâ Êãà¾ã „ÞÞã¦ã½ã ½ãÖ¦Ìã ‡ãŠñ †‡ãŠ †ñÔãñ Ìãõ—ãããä¶ã‡ãŠ ÔãâÔ©ãã¶ã ‡ãŠãè Ô©ãã¹ã¶ãã ãä•ãÔã½ãò ‚㦾ãã£ãìãä¶ã‡ãŠ ‚ã¶ãìÔãâ£ãã¶ã Ôããä֦㠂㣾ãã¹ã¶ã †Ìãâ ãäÍãàã¥ã ‡ãŠã ¹ãî¥ãùã Ôãñ †‡ãŠãè‡ãŠÀ¥ã Öãñý ãä•ã—ããÔãã ¦ã©ãã ÀÞã¶ã㦽ã‡ãŠ¦ãã Ôãñ ¾ãì§ãŠ ÔãÌããó§ã½ã Ôã½ãã‡ãŠÊã¶ã㦽ã‡ãŠ ‚㣾ãã¹ã¶ã ‡ãñŠ ½ã㣾ã½ã Ôãñ ½ããõãäÊã‡ãŠ ãäÌã—ãã¶ã ‡ãŠãñ ÀãñÞã‡ãŠ ºã¶ãã¶ããý ÊãÞããèÊãñ †Ìãâ Ôããè½ããÀãäÖ¦ã / ‚ãÔããè½ã ¹ã㟿ã‰ãŠ½ã ¦ã©ãã ‚ã¶ãìÔãâ£ãã¶ã ¹ããäÀ¾ããñ•ã¶ãã‚ããò ‡ãñŠ ½ã㣾ã½ã Ôãñ œãñ›ãè ‚ãã¾ãì ½ãò Öãè ‚ã¶ãìÔãâ£ãã¶ã àãñ¨ã ½ãò ¹ãÆÌãñÍãý Vision & Mission Establish scientific institution of the highest caliber where teaching and education are totally integrated with state-of-the- art research Make learning of basic sciences exciting through excellent integrative teaching driven by curiosity and creativity Entry into research at an early age through a flexible borderless curriculum and research projects Annual Report 2015-16 Governance Correct Citation Board of Governors IISER Pune Annual Report 2015-16 Chairperson Pune, India Prof. T.V. Ramakrishnan (till 03/12/2015) Emeritus Professor of Physics, DAE Homi Bhabha Professor, Department of Physics, Indian Institute of Science, Bengaluru Published by Dr. K. Venkataramanan (from 04/12/2015) Director and President (Engineering and Construction Projects), Dr. -

Investigating the Co-Regulatory Role of Midline and Extramacrochaetae in Regulating Eye Development and Vision in Drosophila Melanogaster
The University of Southern Mississippi The Aquila Digital Community Honors Theses Honors College Spring 5-2014 Investigating the Co-Regulatory Role of Midline and Extramacrochaetae In Regulating Eye Development and Vision in Drosophila melanogaster Lillian M. Forstall University of Southern Mississippi Follow this and additional works at: https://aquila.usm.edu/honors_theses Part of the Developmental Biology Commons Recommended Citation Forstall, Lillian M., "Investigating the Co-Regulatory Role of Midline and Extramacrochaetae In Regulating Eye Development and Vision in Drosophila melanogaster" (2014). Honors Theses. 236. https://aquila.usm.edu/honors_theses/236 This Honors College Thesis is brought to you for free and open access by the Honors College at The Aquila Digital Community. It has been accepted for inclusion in Honors Theses by an authorized administrator of The Aquila Digital Community. For more information, please contact [email protected]. The University of Southern Mississippi Investigating the Co-Regulatory Role of midline and extramacrochaetae In Regulating Eye Development and Vision in Drosophila melanogaster by Lillian Forstall A Thesis Submitted to the Honors College of The University of Southern Mississippi in Partial Fulfillment of the Requirements for the Degree of Bachelor of Science in the Department of Biological Sciences May 2014 ii Approved by _________________________________ Dr. Sandra Leal, Ph.D., Thesis Adviser Assistant Professor of Biology _________________________________ Dr. Shiao Wang, Ph.D., Interim Chair Department of Biological Sciences _________________________________ Dr. David R. Davies, Ph.D., Dean Honors College iii Abstract The Honors thesis research focused on the roles of extramacrochaetae and midline in regulating eye development and the vision of Drosophila melanogaster. -

The C-Rel Transcription Factor and B-Cell Proliferation: a Deal with the Devil
Oncogene (2004) 23, 2275–2286 & 2004 Nature Publishing Group All rights reserved 0950-9232/04 $25.00 www.nature.com/onc REVIEW The c-Rel transcription factor and B-cell proliferation: a deal with the devil Thomas D Gilmore*,1, Demetrios Kalaitzidis1, Mei-Chih Liang1 and Daniel T Starczynowski1 1Department of Biology, Boston University, 5 Cummington Street, Boston, MA 02215, USA Activation of the Rel/NF-jB signal transduction pathway called the Rel homology domain, which has sequences has been associated with a varietyof animal and human required for DNA binding, dimerization, nuclear malignancies. However, among the Rel/NF-jB family localization, and inhibitor (IkB) binding. Sequences members, onlyc-Rel has been consistentlyshown to be C-terminal to the Rel homology domain in c-Rel, RelA, able to malignantlytransform cells in culture. In addition, and RelB contain transcriptional activation domains, c-rel has been activated bya retroviral promoter insertion and therefore dimers that contain these Rel/NF-kB in an avian B-cell lymphoma, and amplifications of REL members usually increase transcription of target genes. (human c-rel) are frequentlyseen in Hodgkin’s lympho- Although the Rel homology domain sequences of c-Rel mas and diffuse large B-cell lymphomas, and in some proteins across species are highly conserved, their follicular and mediastinal B-cell lymphomas. Phenotypic C-terminal transactivation domains are not. For exam- analysis of c-rel knockout mice demonstrates that c-Rel ple, the Rel homology domains of chicken and human has a normal role in B-cell proliferation and survival; c-Rel are approximately 85% identical, whereas their moreover, c-Rel nuclear activityis required for B-cell C-terminal transactivation domains are only about 10% development.