Supplementary Table 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Overexpresssion of Tyrosine Kinase Discoidin Domain Receptor I (DDR1 ) in Transitional Cell Carcinoma
Overexpresssion of Tyrosine Kinase Discoidin domain receptor I (DDR1 ) in Transitional cell Carcinoma Szu-Ting Chen,* and Shie-Liang Hsieh* Institute and Department of Microbiology and Immunology, National Yang-Ming University, Taipei, Taiwan; To whom proofs are to be sent: Shie-Liang Hsieh, Department and Institute of Microbiology and Immunology, National Yang-Ming University, Shih-Pai, Taipei 112, Taiwan. E-mail address: [email protected] Telephone number: 886-2-28267161 Fax number: 886-2-28277933 INTRODUCTION DDR1, discoidin domain receptor 1, belongs to the novel subfamily of tyrosine kinase receptor, which forms homodimer upon ligand engagement. DDR1 is distinguished from other receptor tyrosine kinase by the discoidin domain in their extracellular domain, which is a homology region originally identified in Dictyostelium discoideum (slime mold) protein, discoidin I, and involves in cells aggregation. Discoidin-1 has binding specificity toward galactose and N-acetyl galactosamine and is essential for slime mold cells adhesion, migration and aggregation during its development, suggesting that DDR1 shared similar biologically function in mammalian [1-3]. DDR1 has been found mainly distributed and in human tissue epithelia, such as kidney, breast, lung [3], bronchial [4] and keratinocytes [5]. Furthermore, DDR1 has been also reported its expression in immune system like the monocyte-derived dendritic cells, annotated as CD167 [6] and macrophage[7]. However, recently, the overexpression of DDR1 has been detected in several human cancers, such as primary breast cancer [1, 8, 9] ovarian[10, 11], brain[12], esophageal cancer [13], and TCC (our data unpublished) in which raising the possibility that DDR1 may play a important role in tumorigenesis [14]. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Defective Lymphocyte Chemotaxis in Я-Arrestin2- and GRK6-Deficient Mice
Defective lymphocyte chemotaxis in -arrestin2- and GRK6-deficient mice Alan M. Fong*, Richard T. Premont*, Ricardo M. Richardson*, Yen-Rei A. Yu†, Robert J. Lefkowitz*‡§, and Dhavalkumar D. Patel*†¶ Departments of *Medicine, ‡Biochemistry, and †Immunology, and §Howard Hughes Medical Institute, Duke University Medical Center, Durham, NC 27710 Contributed by Robert J. Lefkowitz, April 4, 2002 Lymphocyte chemotaxis is a complex process by which cells move kinase, extracellular receptor kinase, and c-jun terminal kinase within tissues and across barriers such as vascular endothelium and activation (9–12), they might also act as positive regulators of is usually stimulated by chemokines such as stromal cell-derived chemotaxis. To evaluate the role of the GRK-arrestin pathway factor-1 (CXCL12) acting via G protein-coupled receptors. Because in chemotaxis, we studied the chemotactic responses of lym- members of this receptor family are regulated (‘‘desensitized’’) by phocytes from -arrestin- and GRK-deficient mice toward G protein-coupled receptor kinase (GRK)-mediated receptor phos- gradients of stromal cell-derived factor 1 (CXCL12), a well  phorylation and -arrestin binding, we examined signaling and characterized chemokine whose receptor is CXCR4, a core- chemotactic responses in splenocytes derived from knockout mice ceptor for HIV. deficient in various -arrestins and GRKs, with the expectation that these responses might be enhanced. Knockouts of -arrestin2, Materials and Methods GRK5, and GRK6 were examined because all three proteins are :expressed at high levels in purified mouse CD3؉ T and B220؉ B Mice. The following mouse strains were used in this study splenocytes. CXCL12 stimulation of membrane GTPase activity was -arrestin2-deficient (back-crossed for six generations onto the unaffected in splenocytes derived from GRK5-deficient mice but C57͞BL6 background; ref. -

Combined Integrin Phosphoproteomic Analyses and Small Interfering RNA–Based Functional Screening Identify Key Regulators for Cancer Cell Adhesion and Migration
Published OnlineFirst April 7, 2009; DOI: 10.1158/0008-5472.CAN-08-2515 Published Online First on April 7, 2009 as 10.1158/0008-5472.CAN-08-2515 Research Article Combined Integrin Phosphoproteomic Analyses and Small Interfering RNA–Based Functional Screening Identify Key Regulators for Cancer Cell Adhesion and Migration Yanling Chen,1 Bingwen Lu,2 Qingkai Yang,1 Colleen Fearns,3 John R. Yates III,2 and Jiing-Dwan Lee1 Departments of 1Immunology, 2Chemical Physiology, and 3Chemistry, The Scripps Research Institute, La Jolla, California Abstract cytoskeletal components (2, 7) and regulation of cell survival, Integrins interact with extracellular matrix (ECM) and deliver proliferation, differentiation, cell cycle, and migration (8, 9). Understanding the mechanism by which integrins modulate these intracellular signaling for cell proliferation, survival, and motility. During tumor metastasis, integrin-mediated cell cellular activities is of significant biological importance. adhesion to and migration on the ECM proteins are required The most common cancers in human include breast cancer, for cancer cell survival and adaptation to the new microen- prostate cancer, lung cancer, colon cancer, and ovarian cancer (10, 11), and their metastasis is the leading cause of mortality in vironment. Using stable isotope labeling by amino acids in cell cancer patients, causing 90% of deaths from solid tumors (11, 12). culture–mass spectrometry, we profiled the phosphoproteo- During the process of metastasis, tumor cells leave the primary mic changes induced by the interactions of cell integrins with site, travel via blood and/or lymphatic circulatory systems, attach type I collagen, the most common ECM substratum. Integrin- to the substratum of ECM at a distant site, and establish a ECM interactions modulate phosphorylation of 517 serine, secondary tumor, accompanied by angiogenesis of the newly threonine, or tyrosine residues in 513 peptides, corresponding formed neoplasm (12). -

Hemoglobin Interaction with Gp1ba Induces Platelet Activation And
ARTICLE Platelet Biology & its Disorders Hemoglobin interaction with GP1bα induces platelet activation and apoptosis: a novel mechanism associated with intravascular hemolysis Rashi Singhal,1,2,* Gowtham K. Annarapu,1,2,* Ankita Pandey,1 Sheetal Chawla,1 Amrita Ojha,1 Avinash Gupta,1 Miguel A. Cruz,3 Tulika Seth4 and Prasenjit Guchhait1 1Disease Biology Laboratory, Regional Centre for Biotechnology, National Capital Region, Biotech Science Cluster, Faridabad, India; 2Biotechnology Department, Manipal University, Manipal, Karnataka, India; 3Thrombosis Research Division, Baylor College of Medicine, Houston, TX, USA, and 4Hematology, All India Institute of Medical Sciences, New Delhi, India *RS and GKA contributed equally to this work. ABSTRACT Intravascular hemolysis increases the risk of hypercoagulation and thrombosis in hemolytic disorders. Our study shows a novel mechanism by which extracellular hemoglobin directly affects platelet activation. The binding of Hb to glycoprotein1bα activates platelets. Lower concentrations of Hb (0.37-3 mM) significantly increase the phos- phorylation of signaling adapter proteins, such as Lyn, PI3K, AKT, and ERK, and promote platelet aggregation in vitro. Higher concentrations of Hb (3-6 mM) activate the pro-apoptotic proteins Bak, Bax, cytochrome c, caspase-9 and caspase-3, and increase platelet clot formation. Increased plasma Hb activates platelets and promotes their apoptosis, and plays a crucial role in the pathogenesis of aggregation and development of the procoagulant state in hemolytic disorders. Furthermore, we show that in patients with paroxysmal nocturnal hemoglobinuria, a chronic hemolytic disease characterized by recurrent events of intravascular thrombosis and thromboembolism, it is the elevated plasma Hb or platelet surface bound Hb that positively correlates with platelet activation. -

Investigating the Role of Cdk11in Animal Cytokinesis
Investigating the Role of CDK11 in Animal Cytokinesis by Thomas Clifford Panagiotou A thesis submitted in conformity with the requirements for the degree of Master of Science Department of Molecular Genetics University of Toronto © Copyright by Thomas Clifford Panagiotou (2020) Investigating the Role of CDK11 in Animal Cytokinesis Thomas Clifford Panagiotou Master of Science Department of Molecular Genetics University of Toronto 2020 Abstract Finely tuned spatio-temporal regulation of cell division is required for genome stability. Cytokinesis constitutes the final stages of cell division, from chromosome segregation to the physical separation of cells, abscission. Abscission is tightly regulated to ensure it occurs after earlier cytokinetic events, like the maturation of the stem body, the regulatory platform for abscission. Active Aurora B kinase enforces the abscission checkpoint, which blocks abscission until chromosomes have been cleared from the cytokinetic machinery. Currently, it is unclear how this checkpoint is overcome. Here, I demonstrate that the cyclin-dependent kinase CDK11 is required for cytokinesis. Both inhibition and depletion of CDK11 block abscission. Furthermore, the mitosis-specific CDK11p58 kinase localizes to the stem body, where its kinase activity rescues the defects of CDK11 depletion and inhibition. These results suggest a model whereby CDK11p58 antagonizes Aurora B kinase to overcome the abscission checkpoint to allow for successful completion of cytokinesis. ii Acknowledgments I am very grateful for the support of my family and friends throughout my studies. I would also like to express my deep gratitude to Wilde Lab members, both past and present, for their advice and collaboration. In particular, I am very grateful to Matthew Renshaw, whose work comprises part of this thesis. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -
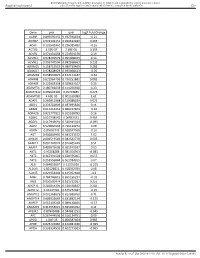
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of