Glucocorticoids Regulate AKR1D1 Activity in Human Liver in Vitro and in Vivo
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification and Developmental Expression of the Full Complement Of
Goldstone et al. BMC Genomics 2010, 11:643 http://www.biomedcentral.com/1471-2164/11/643 RESEARCH ARTICLE Open Access Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish Jared V Goldstone1, Andrew G McArthur2, Akira Kubota1, Juliano Zanette1,3, Thiago Parente1,4, Maria E Jönsson1,5, David R Nelson6, John J Stegeman1* Abstract Background: Increasing use of zebrafish in drug discovery and mechanistic toxicology demands knowledge of cytochrome P450 (CYP) gene regulation and function. CYP enzymes catalyze oxidative transformation leading to activation or inactivation of many endogenous and exogenous chemicals, with consequences for normal physiology and disease processes. Many CYPs potentially have roles in developmental specification, and many chemicals that cause developmental abnormalities are substrates for CYPs. Here we identify and annotate the full suite of CYP genes in zebrafish, compare these to the human CYP gene complement, and determine the expression of CYP genes during normal development. Results: Zebrafish have a total of 94 CYP genes, distributed among 18 gene families found also in mammals. There are 32 genes in CYP families 5 to 51, most of which are direct orthologs of human CYPs that are involved in endogenous functions including synthesis or inactivation of regulatory molecules. The high degree of sequence similarity suggests conservation of enzyme activities for these CYPs, confirmed in reports for some steroidogenic enzymes (e.g. CYP19, aromatase; CYP11A, P450scc; CYP17, steroid 17a-hydroxylase), and the CYP26 retinoic acid hydroxylases. Complexity is much greater in gene families 1, 2, and 3, which include CYPs prominent in metabolism of drugs and pollutants, as well as of endogenous substrates. -

Type of the Paper (Article
Supplementary Material A Proteomics Study on the Mechanism of Nutmeg-induced Hepatotoxicity Wei Xia 1, †, Zhipeng Cao 1, †, Xiaoyu Zhang 1 and Lina Gao 1,* 1 School of Forensic Medicine, China Medical University, Shenyang 110122, P. R. China; lessen- [email protected] (W.X.); [email protected] (Z.C.); [email protected] (X.Z.) † The authors contributed equally to this work. * Correspondence: [email protected] Figure S1. Table S1. Peptide fraction separation liquid chromatography elution gradient table. Time (min) Flow rate (mL/min) Mobile phase A (%) Mobile phase B (%) 0 1 97 3 10 1 95 5 30 1 80 20 48 1 60 40 50 1 50 50 53 1 30 70 54 1 0 100 1 Table 2. Liquid chromatography elution gradient table. Time (min) Flow rate (nL/min) Mobile phase A (%) Mobile phase B (%) 0 600 94 6 2 600 83 17 82 600 60 40 84 600 50 50 85 600 45 55 90 600 0 100 Table S3. The analysis parameter of Proteome Discoverer 2.2. Item Value Type of Quantification Reporter Quantification (TMT) Enzyme Trypsin Max.Missed Cleavage Sites 2 Precursor Mass Tolerance 10 ppm Fragment Mass Tolerance 0.02 Da Dynamic Modification Oxidation/+15.995 Da (M) and TMT /+229.163 Da (K,Y) N-Terminal Modification Acetyl/+42.011 Da (N-Terminal) and TMT /+229.163 Da (N-Terminal) Static Modification Carbamidomethyl/+57.021 Da (C) 2 Table S4. The DEPs between the low-dose group and the control group. Protein Gene Fold Change P value Trend mRNA H2-K1 0.380 0.010 down Glutamine synthetase 0.426 0.022 down Annexin Anxa6 0.447 0.032 down mRNA H2-D1 0.467 0.002 down Ribokinase Rbks 0.487 0.000 -

Hepatic Gene Expression of Bile Acid Synthesis Genes from Wild-Type and Fxr−/− Mice
A 2.0 Acox2 B 2.0 Akr1c14 C 2.0 Akr1d1 D 2.0 Amacr ** 1.5 1.5 1.5 1.5 1.0 1.0 1.0 1.0 0.5 0.5 0.5 0.5 mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA 0.0 0.0 0.0 0.0 GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h FXR WT Fxr−/− FXR WT Fxr−/− FXR WT Fxr−/− FXR WT Fxr−/− E F 2.0 Cyp7b1 2.0 Cyp27a1 G 2.0 Cyp39a1 H 2.0 Hsd3b7 1.5 1.5 1.5 1.5 * 1.0 1.0 1.0 1.0 0.5 0.5 0.5 0.5 mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA 0.0 0.0 0.0 0.0 GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h FXR WT Fxr−/− FXR WT Fxr−/− FXR WT Fxr−/− FXR WT Fxr−/− I J K 2.0 Hsd17b4 2.0 Scp2 2.0 Slc27a5 L Fxr Cyp7a1 Cyp8b1 1.0 1.5 1.5 1.5 * 1.0 1.0 1.0 0.5 *** *** 0.5 0.5 0.5 mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA mRNA (Fold Change) mRNA *** *** *** 0.0 0.0 0.0 0.0 *** GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h GSK − 30’ 1h 2h − 30’ 1h 2h Time (h) 0 4 16 0 4 16 0 4 16 FXR WT Fxr−/− FXR WT Fxr−/− FXR WT Fxr−/− post plating M N CYP7A1 CYP8B1 Supplementary Figure 1 – FXR activation leads to rapid changes in gene expression 1.0 1.0 (A-K) Hepatic gene expression of bile acid synthesis genes from wild-type and Fxr−/− mice. -

TRANSLATIONALLY by AMPK a Dissertation
CHOLESTEROL 7 ALPHA-HYDROXYLASE IS REGULATED POST- TRANSLATIONALLY BY AMPK A dissertation submitted to Kent State University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy By Mauris E.C. Nnamani May 2009 Dissertation written by Mauris E. C. Nnamani B.S, Kent State University, 2006 Ph.D., Kent State University, 2009 Approved by Diane Stroup Advisor Gail Fraizer Members, Doctoral Dissertation Committee S. Vijayaraghavan Arne Gericke Jennifer Marcinkiewicz Accepted by Robert Dorman , Director, School of Biomedical Science John Stalvey , Dean, Collage of Arts and Sciences ii TABLE OF CONTENTS LIST OF FIGURES……………………………………………………………..vi ACKNOWLEDGMENTS……………………………………………………..viii CHAPTER I: INTRODUCTION……………………………………….…........1 a. Bile Acid Synthesis…………………………………………….……….2 i. Importance of Bile Acid Synthesis Pathway………………….….....2 ii. Bile Acid Transport..…………………………………...…...………...3 iii. Bile Acid Synthesis Pathway………………………………………...…4 iv. Classical Bile Acid Synthesis Pathway…..……………………..…..8 Cholesterol 7 -hydroxylase (CYP7A1)……..........………….....8 Transcriptional Regulation of Cholesterol 7 -hydroxylase by Bile Acid-activated FXR…………………………….....…10 CYP7A1 Transcriptional Repression by SHP-dependant Mechanism…………………………………………………...10 CYP7A1 Transcriptional Repression by SHP-independent Mechanism……………………………………..…………….…….11 CYP7A1 Transcriptional Repression by Activated Cellular Kinase…….…………………………...…………………….……12 v. Alternative/ Acidic Bile Acid Synthesis Pathway…………......…….12 Sterol 27-hydroxylase (CYP27A1)……………….…………….12 -

Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2
biology Article Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2 Chiranan Khantham 1 , Wipawadee Yooin 1,2 , Korawan Sringarm 2,3 , Sarana Rose Sommano 2,4 , Supat Jiranusornkul 1, Francisco David Carmona 5,6 , Wutigri Nimlamool 7 , Pensak Jantrawut 1,2, Pornchai Rachtanapun 8,9 and Warintorn Ruksiriwanich 1,2,8,* 1 Department of Pharmaceutical Sciences, Faculty of Pharmacy, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (C.K.); [email protected] (W.Y.); [email protected] (S.J.); [email protected] (P.J.) 2 Cluster of Research and Development of Pharmaceutical and Natural Products Innovation for Human or Animal, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (K.S.); [email protected] (S.R.S.) 3 Department of Animal and Aquatic Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 4 Plant Bioactive Compound Laboratory (BAC), Department of Plant and Soil Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 5 Departamento de Genética e Instituto de Biotecnología, Universidad de Granada, 18071 Granada, Spain; [email protected] 6 Instituto de Investigación Biosanitaria ibs.GRANADA, 18014 Granada, Spain 7 Citation: Khantham, C.; Yooin, W.; Department of Pharmacology, Faculty of Medicine, Chiang Mai University, Chiang Mai 50200, Thailand; Sringarm, K.; Sommano, S.R.; [email protected] 8 Cluster of Agro Bio-Circular-Green Industry, Faculty of Agro-Industry, Chiang Mai University, Jiranusornkul, S.; Carmona, F.D.; Chiang Mai 50100, Thailand; [email protected] Nimlamool, W.; Jantrawut, P.; 9 School of Agro-Industry, Faculty of Agro-Industry, Chiang Mai University, Chiang Mai 50100, Thailand Rachtanapun, P.; Ruksiriwanich, W. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Characterization of Precursor-Dependent Steroidogenesis in Human Prostate Cancer Models
cancers Article Characterization of Precursor-Dependent Steroidogenesis in Human Prostate Cancer Models Subrata Deb 1 , Steven Pham 2, Dong-Sheng Ming 2, Mei Yieng Chin 2, Hans Adomat 2, Antonio Hurtado-Coll 2, Martin E. Gleave 2,3 and Emma S. Tomlinson Guns 2,3,* 1 Department of Pharmaceutical Sciences, College of Pharmacy, Larkin University, Miami, FL 33169, USA; [email protected] 2 The Vancouver Prostate Centre at Vancouver General Hospital, 2660 Oak Street, Vancouver, BC V6H 3Z6, Canada; [email protected] (S.P.); [email protected] (D.-S.M.); [email protected] (M.Y.C.); [email protected] (H.A.); [email protected] (A.H.-C.); [email protected] (M.E.G.) 3 Department of Urologic Sciences, Faculty of Medicine, University of British Columbia, Vancouver, BC V5Z 1M9, Canada * Correspondence: [email protected]; Tel.: +1-604-875-4818 Received: 14 August 2018; Accepted: 17 September 2018; Published: 20 September 2018 Abstract: Castration-resistant prostate tumors acquire the independent capacity to generate androgens by upregulating steroidogenic enzymes or using steroid precursors produced by the adrenal glands for continued growth and sustainability. The formation of steroids was measured by liquid chromatography-mass spectrometry in LNCaP and 22Rv1 prostate cancer cells, and in human prostate tissues, following incubation with steroid precursors (22-OH-cholesterol, pregnenolone, 17-OH-pregnenolone, progesterone, 17-OH-progesterone). Pregnenolone, progesterone, 17-OH-pregnenolone, and 17-OH-progesterone increased C21 steroid (5-pregnan-3,20-dione, 5-pregnan-3,17-diol-20-one, 5-pregnan-3-ol-20-one) formation in the backdoor pathway, and demonstrated a trend of stimulating dihydroepiandrosterone or its precursors in the backdoor pathway in LNCaP and 22Rv1 cells. -

Vitamin D Receptor (VDR) and Cholesterol Homeostasis: Interplay of VDR Enzyme Targets and Vitamin D Deficiency
Vitamin D Receptor (VDR) and Cholesterol Homeostasis: Interplay of VDR Enzyme Targets and Vitamin D Deficiency by Holly P. Quach A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Department of Pharmaceutical Sciences University of Toronto © Copyright by Holly P. Quach, 2016 Vitamin D Receptor (VDR) and Cholesterol Homeostasis: Interplay of VDR Enzyme Targets and Vitamin D Deficiency Holly P. Quach Doctor of Philosophy Department of Pharmaceutical Sciences University of Toronto 2016 Abstract Vitamin D deficiency is speculated to play a role in hypercholesterolemia. However, there has been little molecular evidence to link the two until recent evidence identified the vitamin D receptor (VDR) and its natural ligand, 1α,25-dihydroxyvitamin D3 [1,25(OH)2D3], as key regulators of cholesterol metabolism. In the liver, 1,25(OH)2D3-liganded VDR directly inhibited the small heterodimer partner (Shp) to increase expression of cholesterol 7α-hydroxylase (Cyp7a1), the rate-limiting enzyme for cholesterol metabolism to bile acids, a mechanism independent of the farnesoid X receptor. Vitamin D deficiency was established in mice after 8 weeks of the D-deficient diet, which resulted in decreased levels of plasma and liver 1,25(OH)2D3, downregulation of hepatic Vdr and Cyp7a1, and elevation of Shp. Consequently, higher plasma and liver cholesterol levels were observed. Intervention with 1,25(OH)2D3 or vitamin D3 reversed the altered expression of these cholesterol-regulating genes and lowered cholesterol levels back to baseline levels. The correlations between liver cholesterol vs. liver 1,25(OH)2D3 and Cyp7a1 expression in mice were also found in human liver tissue, suggesting that the VDR could be a ii potential therapeutic target for cholesterol lowering. -

Expression of CYP2S1 in Human Hepatic Stellate Cells
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector FEBS Letters 581 (2007) 781–786 Expression of CYP2S1 in human hepatic stellate cells Carylyn J. Mareka, Steven J. Tuckera, Matthew Korutha, Karen Wallacea,b, Matthew C. Wrighta,b,* a School of Medical Sciences, Institute of Medical Science, University of Aberdeen, Aberdeen, UK b Liver Faculty Research Group, School of Clinical and Laboratory Sciences, University of Newcastle, Newcastle, UK Received 22 November 2006; revised 16 January 2007; accepted 23 January 2007 Available online 2 February 2007 Edited by Laszlo Nagy the expression and accumulation of scarring extracellular Abstract Activated stellate cells are myofibroblast-like cells associated with the generation of fibrotic scaring in chronically fibrotic matrix protein [2]. It is currently thought an inhibition damaged liver. Gene chip analysis was performed on cultured fi- of fibrosis in liver diseases may be an effective approach to brotic stellate cells. Of the 51 human CYP genes known, 13 treating patients for which the cause is refractive to current CYP and 5 CYP reduction-related genes were detected with 4 treatments (e.g. in approx. 30% of hepatitis C infected CYPs (CYP1A1, CYP2E1, CY2S1 and CYP4F3) consistently patients) [2,3]. At present, there is no approved treatment for present in stellate cells isolated from three individuals. Quantita- fibrosis. tive RT-PCR indicated that CYP2S1 was a major expressed Inadvertent toxicity of drugs is often associated with a ‘‘met- CYP mRNA transcript. The presence of a CYP2A-related pro- abolic activation’’ by CYPs [1]. -
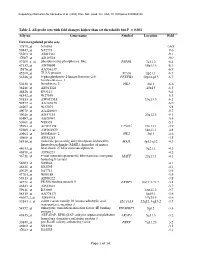
Table 2. All Probe Sets with Fold Changes Higher Than Set Thresholds but P > 0.001 Affy No
Supporting information for Sanoudou et al. (2003) Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0330960100 Table 2. All probe sets with fold changes higher than set thresholds but P > 0.001 Affy no. Gene name Symbol Location Fold Down-regulated probe sets 47879_at N46863 -16.9 59447_at N52773 -9.6 55503_at AI085361 -9.1 47087_at AI310524 -9.0 37209_g_at phosphoserine phosphatase-like PSPHL 7q11.2 -8.4 47137_at AI479899 19p13.3 -8.1 45876_at AA536137 -6.9 45260_at TU3A protein TU3A 3p21.1 -6.7 56246_at 6-phosphofructo-2-kinase/fructose-2,6- PFKFB3 10p14-p15 -6.7 bisphosphatase 3 50230_at hexokinase 2 HK2 2p13 -6.6 38248_at AB011124 20p13 -6.5 44426_at R93141 -6.4 48542_at W27559 -6.3 53813_at AW051518 13q13.3 -6.1 59577_at AA243670 -6.0 46907_at W37075 -5.8 49078_at AA424983 -5.7 49026_at AI357153 20p12.3 -5.4 53487_at AI670947 -5.4 50161_at N39328 -5.1 35994_at AC002398 F25965 19q13.1 -4.9 52285_f_at AW002970 18p11.1 -4.8 40964_at hexokinase 2 HK2 2p13 -4.6 49806_at AI932283 -4.5 58918_at molecule possessing ankyrin repeats induced by MAIL 3p12-q12 -4.3 lipopolysaccharide (MAIL), homolog of mouse 44633_at heat shock 27 kDa associated protein 3q21.1 -4.3 46858_at AI796221 -4.2 36711_at v-maf musculoaponeurotic fibrosarcoma oncogene MAFF 22q13.1 -4.1 homolog F (avian) 54683_at N49844 -4.1 46621_at N32595 -4.1 49629_at N47713 -3.9 47703_at W89189 -3.9 59313_at AI598222 -3.8 34721_at FK506 binding protein 5 FKBP5 6p21.3-21.2 -3.8 46843_at AI632621 -3.7 59611_at R53069 16p11.2 -3.7 58315_at AA778171 3p25.1 -3.6 46607_f_at AI885018 17q25.3 -3.6 33143_s_at solute carrier family 16 (monocarboxylic acid SLC16A3 22q12.3-q13.2 -3.5 transporters), member 3 54152_at eukaryotic translation initiation factor 4E binding EIF4EBP1 8p12 -3.4 protein 1 43935_at ARF-GAP, RHO-GAP, ankyrin repeat and plekstrin ARAP3 5q31.3 -3.2 homology domains-containing protein 3 33849_at pre-B-cell colony-enhancing factor PBEF 7q11.23 -3.2 46902_at N92294 -3.2 47023_at N25555 -3.1 Page 1 of 14 Supporting information for Sanoudou et al. -

Metabolic Enzyme Expression Highlights a Key Role for MTHFD2 and the Mitochondrial Folate Pathway in Cancer
ARTICLE Received 1 Nov 2013 | Accepted 17 Dec 2013 | Published 23 Jan 2014 DOI: 10.1038/ncomms4128 Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer Roland Nilsson1,2,*, Mohit Jain3,4,5,6,*,w, Nikhil Madhusudhan3,4,5, Nina Gustafsson Sheppard1,2, Laura Strittmatter3,4,5, Caroline Kampf7, Jenny Huang8, Anna Asplund7 & Vamsi K. Mootha3,4,5 Metabolic remodeling is now widely regarded as a hallmark of cancer, but it is not clear whether individual metabolic strategies are frequently exploited by many tumours. Here we compare messenger RNA profiles of 1,454 metabolic enzymes across 1,981 tumours spanning 19 cancer types to identify enzymes that are consistently differentially expressed. Our meta- analysis recovers established targets of some of the most widely used chemotherapeutics, including dihydrofolate reductase, thymidylate synthase and ribonucleotide reductase, while also spotlighting new enzymes, such as the mitochondrial proline biosynthetic enzyme PYCR1. The highest scoring pathway is mitochondrial one-carbon metabolism and is centred on MTHFD2. MTHFD2 RNA and protein are markedly elevated in many cancers and correlated with poor survival in breast cancer. MTHFD2 is expressed in the developing embryo, but is absent in most healthy adult tissues, even those that are proliferating. Our study highlights the importance of mitochondrial compartmentalization of one-carbon metabolism in cancer and raises important therapeutic hypotheses. 1 Unit of Computational Medicine, Department of Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 2 Center for Molecular Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 3 Broad Institute, Cambridge, Massachusetts 02142, USA. 4 Department of Systems Biology, Harvard Medical School, Boston, Massachusetts 02115, USA. -

Suppression of Steroid 5Α-Reductase Type I Promotes Cellular Apoptosis
Dou et al. Cell Death and Disease (2021) 12:206 https://doi.org/10.1038/s41419-021-03510-4 Cell Death & Disease ARTICLE Open Access Suppression of steroid 5α-reductase type I promotes cellular apoptosis and autophagy via PI3K/Akt/mTOR pathway in multiple myeloma Renjie Dou1,2, Jinjun Qian2,WeiWu1,YanxinZhang2,YuxiaYuan2, Mengjie Guo1,2,RongfangWei2, Shu Yang2, Artur Jurczyszyn 3, Siegfried Janz 4, Meral Beksac5, Chunyan Gu 1,2 and Ye Yang1,2,6 Abstract Steroid 5α-reductase type I (SRD5A1) is a validated oncogene in many sex hormone-related cancers, but its role in multiple myeloma (MM) remains unknown. Based on gene expression profiling (GEP) of sequential MM samples during the disease course, we found that the aberrant expression of SRD5A1 was correlated with progression and poor prognosis in MM patients. In this study, the oncogenic roles of SRD5A1 were validated in human MM cell lines (ARP1 and H929) and the xenograft MM model as well as the 5TMM mouse model. MTT and flow cytometry were used to assess MM cell proliferation, cell cycle, and apoptosis post inducible knockdown SRD5A1 by lentivirus-mediated short- hairpin RNA (shRNA). Transcriptomic sequencing, immunofluorescence, and western blot were used to investigate the effects of SRD5A1 suppression on cell apoptosis and autophagy. Mechanistically, SRD5A1 downregulation simultaneously regulated both the Bcl-2 family protein-mediated apoptosis and the autophagic process via PI3K/Akt/ mTOR signaling pathway in MM cells. Meanwhile, the autophagy inhibitor (3-methyladenine) and SRD5A1 inhibitor (Dutasteride) were utilized to evaluate their anti-myeloma effect. Thus, our results demonstrated that SRD5A1 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; downregulation simultaneously regulated both the apoptosis and the autophagic process in MM cells.