Molecular Pattern Recognition of A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Quaternary Murid Rodents of Timor Part I: New Material of Coryphomys Buehleri Schaub, 1937, and Description of a Second Species of the Genus
QUATERNARY MURID RODENTS OF TIMOR PART I: NEW MATERIAL OF CORYPHOMYS BUEHLERI SCHAUB, 1937, AND DESCRIPTION OF A SECOND SPECIES OF THE GENUS K. P. APLIN Australian National Wildlife Collection, CSIRO Division of Sustainable Ecosystems, Canberra and Division of Vertebrate Zoology (Mammalogy) American Museum of Natural History ([email protected]) K. M. HELGEN Department of Vertebrate Zoology National Museum of Natural History Smithsonian Institution, Washington and Division of Vertebrate Zoology (Mammalogy) American Museum of Natural History ([email protected]) BULLETIN OF THE AMERICAN MUSEUM OF NATURAL HISTORY Number 341, 80 pp., 21 figures, 4 tables Issued July 21, 2010 Copyright E American Museum of Natural History 2010 ISSN 0003-0090 CONTENTS Abstract.......................................................... 3 Introduction . ...................................................... 3 The environmental context ........................................... 5 Materialsandmethods.............................................. 7 Systematics....................................................... 11 Coryphomys Schaub, 1937 ........................................... 11 Coryphomys buehleri Schaub, 1937 . ................................... 12 Extended description of Coryphomys buehleri............................ 12 Coryphomys musseri, sp.nov.......................................... 25 Description.................................................... 26 Coryphomys, sp.indet.............................................. 34 Discussion . .................................................... -

A Phylogeographic Survey of the Pygmy Mouse Mus Minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome B Sequences of Museum Specimens
A Phylogeographic Survey of the Pygmy Mouse Mus minutoides in South Africa: Taxonomic and Karyotypic Inference from Cytochrome b Sequences of Museum Specimens Pascale Chevret1*, Terence J. Robinson2, Julie Perez3, Fre´de´ric Veyrunes3, Janice Britton-Davidian3 1 Laboratoire de Biome´trie et Biologie Evolutive, UMR CNRS 5558, Universite´ Lyon 1, Villeurbanne, France, 2 Evolutionary Genomics Group, Department of Botany and Zoology, University of Stellenbosch, Stellenbosch, South Africa, 3 Institut des Sciences de l’Evolution de Montpellier, UMR CNRS 5554, Universite´ Montpellier 2, Montpellier, France Abstract The African pygmy mice (Mus, subgenus Nannomys) are a group of small-sized rodents that occur widely throughout sub- Saharan Africa. Chromosomal diversity within this group is extensive and numerous studies have shown the karyotype to be a useful taxonomic marker. This is pertinent to Mus minutoides populations in South Africa where two different cytotypes (2n = 34, 2n = 18) and a modification of the sex determination system (due to the presence of a Y chromosome in some females) have been recorded. This chromosomal diversity is mirrored by mitochondrial DNA sequences that unambiguously discriminate among the various pygmy mouse species and, importantly, the different M. minutoides cytotypes. However, the geographic delimitation and taxonomy of pygmy mice populations in South Africa is poorly understood. To address this, tissue samples of M. minutoides were taken and analysed from specimens housed in six South African museum collections. Partial cytochrome b sequences (400 pb) were successfully amplified from 44% of the 154 samples processed. Two species were identified: M. indutus and M. minutoides. The sequences of the M. indutus samples provided two unexpected features: i) nuclear copies of the cytochrome b gene were detected in many specimens, and ii) the range of this species was found to extend considerably further south than is presently understood. -

Mammals of the Kafa Biosphere Reserve Holger Meinig, Dr Meheretu Yonas, Ondřej Mikula, Mengistu Wale and Abiyu Tadele
NABU’s Follow-up BiodiversityAssessmentBiosphereEthiopia Reserve, Follow-up NABU’s Kafa the at NABU’s Follow-up Biodiversity Assessment at the Kafa Biosphere Reserve, Ethiopia Small- and medium-sized mammals of the Kafa Biosphere Reserve Holger Meinig, Dr Meheretu Yonas, Ondřej Mikula, Mengistu Wale and Abiyu Tadele Table of Contents Small- and medium-sized mammals of the Kafa Biosphere Reserve 130 1. Introduction 132 2. Materials and methods 133 2.1 Study area 133 2.2 Sampling methods 133 2.3 Data analysis 133 3. Results and discussion 134 3.1 Soricomorpha 134 3.2 Rodentia 134 3.3 Records of mammal species other than Soricomorpha or Rodentia 140 4. Evaluation of survey results 143 5. Conclusions and recommendations for conservation and monitoring 143 6. Acknowledgements 143 7. References 144 8. Annex 147 8.1 Tables 147 8.2 Photos 152 NABU’s Follow-up Biodiversity Assessment at the Kafa Biosphere Reserve, Ethiopia Small- and medium-sized mammals of the Kafa Biosphere Reserve Holger Meinig, Dr Meheretu Yonas, Ondřej Mikula, Mengistu Wale and Abiyu Tadele 130 SMALL AND MEDIUM-SIZED MAMMALS Highlights ´ Eight species of rodents and one species of Soricomorpha were found. ´ Five of the rodent species (Tachyoryctes sp.3 sensu (Sumbera et al., 2018)), Lophuromys chrysopus and L. brunneus, Mus (Nannomys) mahomet and Desmomys harringtoni) are Ethiopian endemics. ´ The Ethiopian White-footed Mouse (Stenocephalemys albipes) is nearly endemic; it also occurs in Eritrea. ´ Together with the Ethiopian Vlei Rat (Otomys fortior) and the African Marsh Rat (Dasymys griseifrons) that were collected only during the 2014 survey, seven endemic rodent species are known to occur in the Kafa region, which supports 12% of the known endemic species of the country. -

Rats and Human Health in Africa: Proceedings of an International Workshop on Rodent-Borne Diseases and the Ratzooman Research Project
RatZooMan Workshop 3-6 May 2006, Malelane, Republic of South Africa …………. Rats and Human Health in Africa: Proceedings of an international workshop on rodent-borne diseases and the RatZooMan research project 3-6 May 2006, The Pestana Kruger Lodge, Malelane, Mpumalanga Province, Republic of South Africa Page 1 RatZooMan Workshop 3-6 May 2006, Malelane, Republic of South Africa …………. This is an output from EC INCO-DEV FP5 research contract: ICA4 CT2002 10056, The prevention of sanitary risks linked to rodents at the rural/peri-urban interface, ratzooman http://www.nri.org/ratzooman © University of Greenwich 2006 The Natural Resources Institute of the University of Greenwich is an internationally recognized centre of expertise in research and consultancy in the environment and natural resources sector. The Institute carious out research, development and training to promote efficient management and use of renewable natural resources in support of sustainable livelihoods. Short extracts of material from this publication may be reproduced in any non-advertising, non-profit- making context provided that the source is acknowledged. Permission for commercial reproduction should be sought from the Managing Editor, Natural Resources Institute, Central Avenue, Chatham Maritime, Kent ME4 4TB, United Kingdom Page 2 RatZooMan Workshop 3-6 May 2006, Malelane, Republic of South Africa …………. Table of Contents Preface....................................................................................................................... 4 Summary................................................................................................................... -

Evolutionary Characterization of a Y Chromosomal Sequence Conserved in the Genus Mus
Genet. Res., Camb. (1988), 52, pp. 145-150 With 5 text-figures Printed in Great Britain 145 Evolutionary characterization of a Y chromosomal sequence conserved in the genus Mus YUTAKA NISHIOKA Department of Biology, McGill University, Montreal, Quebec, Canada H3A IBI (Received 8 October 1987 and in revised form 26 January 1988) Summary The extent of accumulation of mouse Y chromosomal repetitive sequences generally correlates with the known phylogenetic relationships in the genus Mus. However, we describe here a M. musculus Y chromosomal repetitive sequence, designated as ACClfl, whose accumulation patterns among eight Mus species do not correspond to their phylogenetic relationships. Although male-specific hybridization bands were present in all the species examined, significant accumulation (^ 200 copies) in the Y chromosomes was found in M. minutoides (subgenus Nannomys), M. pahari (subgenus Coelomys) and M. saxicola (subgenus Pyromys) as well as in the three closely related species M. hortulanus, M. musculus and M. spretus that belong to the subgenus Mus. Unexpectedly, the Y chromosomes of M. caroli and M. cookii (both subgenus Mus) had considerably reduced amounts of ACClfl-related sequences. Furthermore, in rats {Rattus norvegicus) the major accumulation sites appear to be autosomal. These observations suggest that caution must be taken in the interpretation of data obtained with repetitive sequences that have evolved quickly. 1. Introduction over 50 mouse (M. musculus) Y chromosomal sequences (Nishioka & Lamothe, 1987a). To date, 32 Recently several groups have isolated mouse DNA fragments were generated from 11 original (Mus musculus) Y chromosomal repetitive sequences isolates and their conservation in the genus Mus has (Bishop et al. -

Increased Geographic Sampling Reveals Considerable New Genetic
Mammalian Biology 79 (2014) 24–35 Contents lists available at ScienceDirect Mammalian Biology jou rnal homepage: www.elsevier.com/locate/mambio Original Investigation Increased geographic sampling reveals considerable new genetic diversity in the morphologically conservative African Pygmy Mice (Genus Mus; Subgenus Nannomys) a,∗ a d a,b,c Jennifer Lamb , Sarah Downs , Seth Eiseb , Peter John Taylor a School of Life Sciences, New Biology Building, University of KwaZulu-Natal, University Road, Westville, KwaZulu-Natal 3630, South Africa b Department of Ecology and Resource Management, School of Environmental Sciences, University of Venda, Post Bag X5050, Thohoyandou 0950, South Africa c Core Team Member, Centre for Invasion Biology, Department of Botany and Zoology, Stellenbosch University, Post Bag X1, Matieland 7602, South Africa d University of Namibia, Windhoek, Namibia a r a t i b s c l e i n f o t r a c t Article history: African endemic pygmy mice (Genus Mus; sub-genus Nannomys) have considerable economic and public Received 7 March 2013 health significance, and some species exhibit novel sex determination systems, making accurate knowl- Accepted 19 August 2013 edge of their phylogenetics and distribution limits important. This phylogenetic study was based on the by Frank E. Zachos mitochondrial control region and cytochrome b gene, for which a substantial body of published data was Available online 13 September 2013 available. Study specimens were sourced from eight previously unsampled or poorly sampled countries, and include samples morphologically identified as Mus bufo, M. indutus, M. callewaerti, M. triton and M. Keywords: neavei. These analyses increase the known genetic diversity of Nannomys from 65 to 102 haplotypes; at Nannomys least 5 unassigned haplotypes are distinguished by potentially species-level cytochrome b genetic dis- Mus bufo tances. -

The Testis-Determining Gene, SRY, Exists in Multiple Copies in Old World Rodents
Genet. Res., Camb. (1994), 64, pp. 151-159 With 6 text-figures Copyright © 1994 Printed in Great Britain 151 The testis-determining gene, SRY, exists in multiple copies in Old World rodents CLAUDE M. NAGAMINE Department of Cell Biology, Vanderbilt University School of Medicine, Medical Center North, Room T2216, Nashville, TN USA 37232-2175 (Received 19 July 1994) Summary SR Y is a unique gene on the Y chromosome in most mammalian species including the laboratory mouse, Mus musculus, and the closely related European wild mouse species M. spicilegus, M. macedonicus, and M. spretus. In contrast, SR Y is present in 2-6 copies in the more distantly related Asian mouse species M. caroli, M. cervicolor, and M. cookii and in 2—13 copies in the related murid species Pyromys saxicola, Coelomys pahari, Nannomys minutoides, Mastomys natalensis, and Rattus norvegicus. Copy numbers do not correlate with known phylogenetic relationships suggesting that SR Y has undergone a rapid and complex evolution in these species. SRY was recently proposed as a molecular probe for phylogenetic inferences. The presence of multiple SR Y genes in a wide range of murid species and genera, and at least one cricetid species, necessitates caution in the use of SR Y for phylogenetic studies in the Rodentia unless it is ascertained that multiple SR Y genes do not exist. family whose signature or characteristic amino acid 1. Introduction pattern is a DNA-binding domain of approximately Mammalian sex determination pivots on the absence 85 amino acids designated the HMG domain. The or presence of a Y chromosome. -

The Involvement of Epithelial Cells in Arenavirus-Induced Pathogenesis
University of Louisville ThinkIR: The University of Louisville's Institutional Repository Electronic Theses and Dissertations 5-2018 The involvement of epithelial cells in arenavirus-induced pathogenesis. Nikole Leslie Margaret Warner University of Louisville Follow this and additional works at: https://ir.library.louisville.edu/etd Part of the Virology Commons Recommended Citation Warner, Nikole Leslie Margaret, "The involvement of epithelial cells in arenavirus-induced pathogenesis." (2018). Electronic Theses and Dissertations. Paper 2964. https://doi.org/10.18297/etd/2964 This Doctoral Dissertation is brought to you for free and open access by ThinkIR: The University of Louisville's Institutional Repository. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of ThinkIR: The University of Louisville's Institutional Repository. This title appears here courtesy of the author, who has retained all other copyrights. For more information, please contact [email protected]. THE INVOLVEMENT OF EPITHELIAL CELLS IN ARENAVIRUS-INDUCED PATHOGENESIS By Nikole Leslie Margaret Warner B.S., Marian University, 2010 M.S., University of Louisville, 2014 A Dissertation Submitted to the Faculty of the School of Medicine of the University of Louisville In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Microbiology and Immunology Department of Microbiology and Immunology University of Louisville Louisville, KY May 2018 Copyright 2018 by Nikole Leslie Margaret Warner All rights reserved THE INVOLVEMENT OF EPITHELIAL CELLS IN ARENAVIRUS-INDUCED PATHOGENESIS By Nikole Leslie Margaret Warner B.S., Marian University, 2010 M.S., University of Louisville, 2014 A Dissertation Approved on April 16, 2018 By the Following Dissertation Committee Members: Igor Lukashevich, Ph.D. -
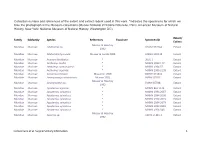
Collection Number and References of the Extant and Extinct Rodent Used in This Work
Collection number and references of the extant and extinct rodent used in this work. *indicates the specimens for which we take the photograph in the Museum collections (Musée National d’Histoire Naturelle, Paris; American Museum of Natural History, New York; National Museum of Natural History, Washington DC). Extant/ Family Subfamily Species References Fossil site Specimen ID Extinct Musser & Headley Muridae Murinae Abditomys sp. USNM 357244 Extant 1992 Muridae Murinae Abelomelomys sevia Musser & Lunde 2009 AMNH 192119 Extant Muridae Murinae Acomys dimidiatus * 2001 3 Extant Muridae Murinae Aethomys hindei * MNHN 1999 177 Extant Muridae Murinae Aethomys namanquesis * MNHN 1964 57 Extant Muridae Murinae Aethomys nigeriae * MNHN 1996 2239 Extant Muridae Murinae Anisomys imitator Missonne 1969 BMNH 471310 Extant Muridae Murinae Anonymomys mindorensis Musser 1981 FMNH 87597 Extant Musser & Headley Muridae Murinae Anonymomys sp. FMNH 87598 Extant 1992 Muridae Murinae Apodemus agrarius * MNHN BL6 1134 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2667 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2668 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2671 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2679 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 2681 Extant Muridae Murinae Apodemus sylvaticus * MNHN 1994 945 Extant Musser & Headley Muridae Murinae Apomys sp. 12971 CLM1-3 Extant 1992 Gomez Cano et al. Supplementary Information 1 Collection number and references of the extant and extinct rodent used in this work. *indicates the specimens for which we take the photograph in the Museum collections (Musée National d’Histoire Naturelle, Paris; American Museum of Natural History, New York; National Museum of Natural History, Washington DC). Extant/ Family Subfamily Species References Fossil site Specimen ID Extinct Musser & Headley Muridae Murinae Archboldomys sp. -

Novel Arenavirus Sequences in Hylomyscus Sp. and Mus (Nannomys) Setulosus from Coˆte D’Ivoire: Implications for Evolution of Arenaviruses in Africa
Novel Arenavirus Sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Coˆte d’Ivoire: Implications for Evolution of Arenaviruses in Africa David Coulibaly-N’Golo1,2., Bernard Allali1., Ste´phane K. Kouassi1, Elisabeth Fichet-Calvet3, Beate Becker-Ziaja2, Toni Rieger2, Stephan O¨ lschla¨ger2, Hernri Dosso4, Christiane Denys3, Jan ter Meulen5,6, Chantal Akoua-Koffi1, Stephan Gu¨ nther2* 1 Laboratoire des Arbovirus/Ente´rovirus, Institut Pasteur de Coˆte d’Ivoire, Abidjan, Coˆte d’Ivoire, 2 Department of Virology, Bernhard-Nocht-Institute for Tropical Medicine, Hamburg, Germany, 3 De´partement Syste´matique et Evolution, Museum National d’Histoire Naturelle, Paris, France, 4 Centre de Recherche en Ecologie, Universite d’Abobo-Adjame, Abidjan, Coˆte d’Ivoire, 5 Institute of Virology, Philipps University, Marburg, Germany, 6 Merck Research Laboratories, West Point, Pennsylvania, United States of America Abstract This study aimed to identify new arenaviruses and gather insights in the evolution of arenaviruses in Africa. During 2003 through 2005, 1,228 small mammals representing 14 different genera were trapped in 9 villages in south, east, and middle west of Coˆte d’Ivoire. Specimens were screened by pan-Old World arenavirus RT-PCRs targeting S and L RNA segments as well as immunofluorescence assay. Sequences of two novel tentative species of the family Arenaviridae, Menekre and Gbagroube virus, were detected in Hylomyscus sp. and Mus (Nannomys) setulosus, respectively. Arenavirus infection of Mus (Nannomys) setulosus was also demonstrated by serological testing. Lassa virus was not found, although 60% of the captured animals were Mastomys natalensis. Complete S RNA and partial L RNA sequences of the novel viruses were recovered from the rodent specimens and subjected to phylogenetic analysis. -

Desert Pygmy Mouse
Mus indutus – Desert Pygmy Mouse Assessment Rationale Listed as Least Concern because it ranges extensively within the assessment region and at times has been recorded as extremely abundant due to occasional population explosions. The rate of habitat loss in the North West Province, which constitutes a large portion of the species’ extent of occurrence, was 12% between 1994 and 2010. However, because this species is able to exist in agricultural habitats and wildlife ranching is expanding in the North West Province, we retain the Least Concern listing. Due to confusion with M. minutoides in the past, Richard Yarnell the distribution of this species has not been resolved. Further vetting of museum records and field surveys to Regional Red List status (2016) Least Concern determine occupancy are required in order to resolve the uncertainty around this species. National Red List status (2004) Least Concern Regional population effects: There is presumably Reasons for change No change dispersal across contiguous habitat from Botswana and Global Red List status (2016) Least Concern Namibia into Kgalagadi Transfrontier Park and surrounding areas. TOPS listing (NEMBA) (2007) None CITES listing None Distribution Endemic No The geographic extent of this species’ range is largely uncertain, owing to the confusion with M. minutoides The Desert Pygmy Mouse is so named due to its (Lamb et al. 2014). However, based on molecular extensive range through the semi-arid terrain of grounds, it has been confirmed from three widely South Africa and Botswana. separated localities across South Africa and Botswana, suggesting that it has a wide distribution across the arid and semi-arid regions of southern Africa (Lamb et al. -

Aardvark 25, 53 Abalosia 146 Abrocoma 141, 386 Abrocomidae 29
Cambridge University Press 978-1-107-04433-3 -Evolution of the Rodents: Advances in Phylogeny, Functional Morphology and Development Edited by Philip G. Cox and Lionel Hautier Index More information Index aardvark 25, 53 Alagomyidae 74–5 Abalosia 146 Alagomyidea 75, 81 Abrocoma 141, 386 Alagomys 73–5 Abrocomidae 29, 141, 283, 483 dentition 75 Abudhabia 195 Alagomys inopinatus 74 Acarechimys 146 Alagomys oriensis 74 Acaremyidae 406 Alagomys russelli 74 Acaremys 146 Allactaga 388, 397 Acomys 188–91, 193, 203, 391 Allactaginae 43, 388 Acomys cahirinus 203 allometry 235, 285, 287, 311, 314, 533 Acomys chrysophilus 203 and molars 315 Acomys coppensi 192 Allophaiomys 495, 498 Acomys ineptus 203 Allotheria 324 Acomys kaiseri 203 Alphagaulus 431 Acomys lavocati 203 Anagalida 5–6 Acomys spinosissimus 190–1 anagenesis 140, 150, 154–6 Acomys subspinosus 190 Andinomys 466 Aconaemys porteri 144 annular ligament 382, 389 Aconaemys sagei 144 anoles 57 acouchis 29 Anomaluridae 26, 43, 81, 387–8, 435, 483 acoustic impedance 394 Anomaluroidea 21, 71, 378, 383, 387, 393 acoustic stiffness 394 middle ear 387 Actenomys 148 origin and dispersal 100 Actenomys priscus 144, 150 ossicles 392 adaptive radiation 55–6, 100, 202, 277, 539 Anomaluromorpha 26–8, 43, 290–1, 426, 483 Palaeogene Asian radiation 98 Anomalurus 26, 388 placental 56 anteaters 25, 53 adelomyine 393 Antemus 51, 193, 195 Adelphomys 145 Antemus chinjiensis 192–3, 196 Adu Asa (fossil site) 199 Antemus mancharensis 51, 192–3 Aeretes 223 Anthropoidea 101, 328 Aeromys thomasi 338–9 Apatemyidae 3 Aethomys