Loss of Wave1 Gene Defines a Subtype of Lethal Prostate Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genome-Wide Analysis of Host-Chromosome Binding Sites For
Lu et al. Virology Journal 2010, 7:262 http://www.virologyj.com/content/7/1/262 RESEARCH Open Access Genome-wide analysis of host-chromosome binding sites for Epstein-Barr Virus Nuclear Antigen 1 (EBNA1) Fang Lu1, Priyankara Wikramasinghe1, Julie Norseen1,2, Kevin Tsai1, Pu Wang1, Louise Showe1, Ramana V Davuluri1, Paul M Lieberman1* Abstract The Epstein-Barr Virus (EBV) Nuclear Antigen 1 (EBNA1) protein is required for the establishment of EBV latent infection in proliferating B-lymphocytes. EBNA1 is a multifunctional DNA-binding protein that stimulates DNA replication at the viral origin of plasmid replication (OriP), regulates transcription of viral and cellular genes, and tethers the viral episome to the cellular chromosome. EBNA1 also provides a survival function to B-lymphocytes, potentially through its ability to alter cellular gene expression. To better understand these various functions of EBNA1, we performed a genome-wide analysis of the viral and cellular DNA sites associated with EBNA1 protein in a latently infected Burkitt lymphoma B-cell line. Chromatin-immunoprecipitation (ChIP) combined with massively parallel deep-sequencing (ChIP-Seq) was used to identify cellular sites bound by EBNA1. Sites identified by ChIP- Seq were validated by conventional real-time PCR, and ChIP-Seq provided quantitative, high-resolution detection of the known EBNA1 binding sites on the EBV genome at OriP and Qp. We identified at least one cluster of unusually high-affinity EBNA1 binding sites on chromosome 11, between the divergent FAM55 D and FAM55B genes. A con- sensus for all cellular EBNA1 binding sites is distinct from those derived from the known viral binding sites, sug- gesting that some of these sites are indirectly bound by EBNA1. -

UCSD MOLECULE PAGES Doi:10.6072/H0.MP.A002549.01 Volume 1, Issue 2, 2012 Copyright UC Press, All Rights Reserved
UCSD MOLECULE PAGES doi:10.6072/H0.MP.A002549.01 Volume 1, Issue 2, 2012 Copyright UC Press, All rights reserved. Review Article Open Access WAVE2 Tadaomi Takenawa1, Shiro Suetsugu2, Daisuke Yamazaki3, Shusaku Kurisu1 WASP family verprolin-homologous protein 2 (WAVE2, also called WASF2) was originally identified by its sequence similarity at the carboxy-terminal VCA (verprolin, cofilin/central, acidic) domain with Wiskott-Aldrich syndrome protein (WASP) and N-WASP (neural WASP). In mammals, WAVE2 is ubiquitously expressed, and its two paralogs, WAVE1 (also called suppressor of cAMP receptor 1, SCAR1) and WAVE3, are predominantly expressed in the brain. The VCA domain of WASP and WAVE family proteins can activate the actin-related protein 2/3 (Arp2/3) complex, a major actin nucleator in cells. Proteins that can activate the Arp2/3 complex are now collectively known as nucleation-promoting factors (NPFs), and the WASP and WAVE families are a founding class of NPFs. The WAVE family has an amino-terminal WAVE homology domain (WHD domain, also called the SCAR homology domain, SHD) followed by the proline-rich region that interacts with various Src-homology 3 (SH3) domain proteins. The VCA domain located at the C-terminus. WAVE2, like WAVE1 and WAVE3, constitutively forms a huge heteropentameric protein complex (the WANP complex), binding through its WHD domain with Abi-1 (or its paralogs, Abi-2 and Abi-3), HSPC300 (also called Brick1), Nap1 (also called Hem-2 and NCKAP1), Sra1 (also called p140Sra1 and CYFIP1; its paralog is PIR121 or CYFIP2). The WANP complex is recruited to the plasma membrane by cooperative action of activated Rac GTPases and acidic phosphoinositides. -

Defining Functional Interactions During Biogenesis of Epithelial Junctions
ARTICLE Received 11 Dec 2015 | Accepted 13 Oct 2016 | Published 6 Dec 2016 | Updated 5 Jan 2017 DOI: 10.1038/ncomms13542 OPEN Defining functional interactions during biogenesis of epithelial junctions J.C. Erasmus1,*, S. Bruche1,*,w, L. Pizarro1,2,*, N. Maimari1,3,*, T. Poggioli1,w, C. Tomlinson4,J.Lees5, I. Zalivina1,w, A. Wheeler1,w, A. Alberts6, A. Russo2 & V.M.M. Braga1 In spite of extensive recent progress, a comprehensive understanding of how actin cytoskeleton remodelling supports stable junctions remains to be established. Here we design a platform that integrates actin functions with optimized phenotypic clustering and identify new cytoskeletal proteins, their functional hierarchy and pathways that modulate E-cadherin adhesion. Depletion of EEF1A, an actin bundling protein, increases E-cadherin levels at junctions without a corresponding reinforcement of cell–cell contacts. This unexpected result reflects a more dynamic and mobile junctional actin in EEF1A-depleted cells. A partner for EEF1A in cadherin contact maintenance is the formin DIAPH2, which interacts with EEF1A. In contrast, depletion of either the endocytic regulator TRIP10 or the Rho GTPase activator VAV2 reduces E-cadherin levels at junctions. TRIP10 binds to and requires VAV2 function for its junctional localization. Overall, we present new conceptual insights on junction stabilization, which integrate known and novel pathways with impact for epithelial morphogenesis, homeostasis and diseases. 1 National Heart and Lung Institute, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. 2 Computing Department, Imperial College London, London SW7 2AZ, UK. 3 Bioengineering Department, Faculty of Engineering, Imperial College London, London SW7 2AZ, UK. 4 Department of Surgery & Cancer, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. -

Systems Analysis Implicates WAVE2&Nbsp
JACC: BASIC TO TRANSLATIONAL SCIENCE VOL.5,NO.4,2020 ª 2020 THE AUTHORS. PUBLISHED BY ELSEVIER ON BEHALF OF THE AMERICAN COLLEGE OF CARDIOLOGY FOUNDATION. THIS IS AN OPEN ACCESS ARTICLE UNDER THE CC BY-NC-ND LICENSE (http://creativecommons.org/licenses/by-nc-nd/4.0/). PRECLINICAL RESEARCH Systems Analysis Implicates WAVE2 Complex in the Pathogenesis of Developmental Left-Sided Obstructive Heart Defects a b b b Jonathan J. Edwards, MD, Andrew D. Rouillard, PHD, Nicolas F. Fernandez, PHD, Zichen Wang, PHD, b c d d Alexander Lachmann, PHD, Sunita S. Shankaran, PHD, Brent W. Bisgrove, PHD, Bradley Demarest, MS, e f g h Nahid Turan, PHD, Deepak Srivastava, MD, Daniel Bernstein, MD, John Deanfield, MD, h i j k Alessandro Giardini, MD, PHD, George Porter, MD, PHD, Richard Kim, MD, Amy E. Roberts, MD, k l m m,n Jane W. Newburger, MD, MPH, Elizabeth Goldmuntz, MD, Martina Brueckner, MD, Richard P. Lifton, MD, PHD, o,p,q r,s t d Christine E. Seidman, MD, Wendy K. Chung, MD, PHD, Martin Tristani-Firouzi, MD, H. Joseph Yost, PHD, b u,v Avi Ma’ayan, PHD, Bruce D. Gelb, MD VISUAL ABSTRACT Edwards, J.J. et al. J Am Coll Cardiol Basic Trans Science. 2020;5(4):376–86. ISSN 2452-302X https://doi.org/10.1016/j.jacbts.2020.01.012 JACC: BASIC TO TRANSLATIONALSCIENCEVOL.5,NO.4,2020 Edwards et al. 377 APRIL 2020:376– 86 WAVE2 Complex in LVOTO HIGHLIGHTS ABBREVIATIONS AND ACRONYMS Combining CHD phenotype–driven gene set enrichment and CRISPR knockdown screening in zebrafish is an effective approach to identifying novel CHD genes. -

Supporting Information
Supporting Information Edgar et al. 10.1073/pnas.1601895113 SI Methods (Actimetrics), and recordings were analyzed using LumiCycle Mice. Sample size was determined using the resource equation: Data Analysis software (Actimetrics). E (degrees of freedom in ANOVA) = (total number of exper- – Cell Cycle Analysis of Confluent Cell Monolayers. NIH 3T3, primary imental animals) (number of experimental groups), with −/− sample size adhering to the condition 10 < E < 20. For com- WT, and Bmal1 fibroblasts were sequentially transduced − − parison of MuHV-4 and HSV-1 infection in WT vs. Bmal1 / with lentiviral fluorescent ubiquitin-based cell cycle indicators mice at ZT7 (Fig. 2), the investigator did not know the genotype (FUCCI) mCherry::Cdt1 and amCyan::Geminin reporters (32). of the animals when conducting infections, bioluminescence Dual reporter-positive cells were selected by FACS (Influx Cell imaging, and quantification. For bioluminescence imaging, Sorter; BD Biosciences) and seeded onto 35-mm dishes for mice were injected intraperitoneally with endotoxin-free lucif- subsequent analysis. To confirm that expression of mCherry:: Cdt1 and amCyan::Geminin correspond to G1 (2n DNA con- erin (Promega E6552) using 2 mg total per mouse. Following < ≤ anesthesia with isofluorane, they were scanned with an IVIS tent) and S/G2 (2 n 4 DNA content) cell cycle phases, Lumina (Caliper Life Sciences), 15 min after luciferin admin- respectively, cells were stained with DNA dye DRAQ5 (abcam) and analyzed by flow cytometry (LSR-Fortessa; BD Biosci- istration. Signal intensity was quantified using Living Image ences). To examine dynamics of replicative activity under ex- software (Caliper Life Sciences), obtaining maximum radiance perimental confluent conditions, synchronized FUCCI reporter for designated regions of interest (photons per second per − − − monolayers were observed by time-lapse live cell imaging over square centimeter per Steradian: photons·s 1·cm 2·sr 1), relative 3 d (Nikon Eclipse Ti-E inverted epifluorescent microscope). -

Effects and Mechanisms of Eps8 on the Biological Behaviour of Malignant Tumours (Review)
824 ONCOLOGY REPORTS 45: 824-834, 2021 Effects and mechanisms of Eps8 on the biological behaviour of malignant tumours (Review) KAILI LUO1, LEI ZHANG2, YUAN LIAO1, HONGYU ZHOU1, HONGYING YANG2, MIN LUO1 and CHEN QING1 1School of Pharmaceutical Sciences and Yunnan Key Laboratory of Pharmacology for Natural Products, Kunming Medical University, Kunming, Yunnan 650500; 2Department of Gynecology, Yunnan Tumor Hospital and The Third Affiliated Hospital of Kunming Medical University; Kunming, Yunnan 650118, P.R. China Received August 29, 2020; Accepted December 9, 2020 DOI: 10.3892/or.2021.7927 Abstract. Epidermal growth factor receptor pathway substrate 8 1. Introduction (Eps8) was initially identified as the substrate for the kinase activity of EGFR, improving the responsiveness of EGF, which Malignant tumours are uncontrolled cell proliferation diseases is involved in cell mitosis, differentiation and other physiological caused by oncogenes and ultimately lead to organ and body functions. Numerous studies over the last decade have demon- dysfunction (1). In recent decades, great progress has been strated that Eps8 is overexpressed in most ubiquitous malignant made in the study of genes and signalling pathways in tumours and subsequently binds with its receptor to activate tumorigenesis. Eps8 was identified by Fazioli et al in NIH-3T3 multiple signalling pathways. Eps8 not only participates in the murine fibroblasts via an approach that allows direct cloning regulation of malignant phenotypes, such as tumour proliferation, of intracellular substrates for receptor tyrosine kinases (RTKs) invasion, metastasis and drug resistance, but is also related to that was designed to study the EGFR signalling pathway. Eps8 the clinicopathological characteristics and prognosis of patients. -

Identification of Novel E2F1 Target Genes Regulated in Cell Cycle
Oncogene (2006) 25, 1786–1798 & 2006 Nature Publishing Group All rights reserved 0950-9232/06 $30.00 www.nature.com/onc ORIGINAL ARTICLE Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners R Iwanaga1,3, H Komori1, S Ishida2, N Okamura3, K Nakayama4, KI Nakayama5 and K Ohtani1 1Human Gene Sciences Center, Tokyo Medical and Dental University, Bunkyo-ku, Tokyo, Japan; 2Division of Pharmacology, National Institute of Health Sciences, Setagaya-ku, Tokyo, Japan; 3Laboratory of Microbiology and Immunology, Graduate School of Health Sciences, Tokyo Medical and Dental University, Bunkyo-ku, Tokyo, Japan; 4Department of Developmental Biology, Center for Translational and Advanced Animal Research on Human Disease, Graduate School of Medicine, Tohoku University, Aoba-ku, Sendai, Japan and 5Department of Molecular and Cellular Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka, Japan The transcription factor E2F mediates cell cycle-depen- cell cycle progression (Nevins et al., 1997; Dyson, 1998; dent expression of genes important for cell proliferation in Trimarchi and Lees, 2002). The transcriptional ability of response to growth stimulation. To further understand the E2F is cell cycle-regulated mainly through association role of E2F, we utilized a sensitive subtraction method to with the retinoblastoma tumor suppressor family of explore new E2F1 targets, which are expressed at low proteins pRb, p107 and p130. During the progression of levels and might have been unrecognized in previous cells from G1 to S phase, G1 cyclin-dependent kinases studies. We identified 33 new E2F1-inducible genes, (cdks)phosphorylate and dissociate the pRb family including checkpoint genes Claspin and Rad51ap1, and proteins from E2F, resulting in the activation of a group four genes with unknown function required for cell cycle of genes required for progression into the S phase. -

Mouse Wasf1 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Wasf1 Knockout Project (CRISPR/Cas9) Objective: To create a Wasf1 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Wasf1 gene (NCBI Reference Sequence: NM_031877 ; Ensembl: ENSMUSG00000019831 ) is located on Mouse chromosome 10. 9 exons are identified, with the ATG start codon in exon 2 and the TAA stop codon in exon 9 (Transcript: ENSMUST00000019975). Exon 3~7 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mutation of this gene has been associated with both morphological and functional defects of the central nervous system. Targeted mutagenesis has resulted in mice that display sensorimotor and cognitive defects similar to those exhibited by patients with 3p-syndrome mental retardation. Exon 3 starts from about 7.99% of the coding region. Exon 3~7 covers 45.32% of the coding region. The size of effective KO region: ~8173 bp. The KO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 3 4 5 6 7 9 Legends Exon of mouse Wasf1 Knockout region Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 3 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. -

Identification of Potential Key Genes and Pathway Linked with Sporadic Creutzfeldt-Jakob Disease Based on Integrated Bioinformatics Analyses
medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Identification of potential key genes and pathway linked with sporadic Creutzfeldt-Jakob disease based on integrated bioinformatics analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Abstract Sporadic Creutzfeldt-Jakob disease (sCJD) is neurodegenerative disease also called prion disease linked with poor prognosis. The aim of the current study was to illuminate the underlying molecular mechanisms of sCJD. The mRNA microarray dataset GSE124571 was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were screened. -

Analysis of the Complex Interaction of Cdr1as‑Mirna‑Protein and Detection of Its Novel Role in Melanoma
ONCOLOGY LETTERS 16: 1219-1225, 2018 Analysis of the complex interaction of CDR1as‑miRNA‑protein and detection of its novel role in melanoma LIHUAN ZHANG*, YUAN LI*, WENYAN LIU, HUIFENG LI and ZHIWEI ZHU College of Life Sciences, Shanxi Agricultural University, Taigu, Shanxi 030801, P.R. China Received May 15, 2017; Accepted April 9, 2018 DOI: 10.3892/ol.2018.8700 Abstract. Despite improvements in the prevention, diagnosis has been detected in several species, including viruses (2), and treatment of melanoma having developed rapidly, the role plants (4), archaea (5)[Salzman, 2013 #198; Memczak, 2013 of circular RNA CDR1 antisense RNA (CDR1as) in melanoma #291] and animals (6). In eukaryotic cells, circRNA has remains to be elucidated. The aim of the present study was to attracted attention due to its unique characteristics, including predict the novel roles of CDR1as in melanoma through novel high stability, specificity and evolutionary conservation (7). bioinformatics analysis. In the present study, the circ2Traits The majority of circRNAs are derived from exons of coding database was used to supply information on CDR1as in cancer. regions, 3'UTRs, 5'UTRs, introns, intergenetic regions and CircNet, circBase and circInteractome databases were used to antisense RNAs (8). CircRNAs, which have attracted attention detect the co-expression of CDR1as, microRNAs and proteins. in recent years, can be produced by canonical and nonca- Furthermore, the functions and pathways of the associated nonical splicing as distinguished from the linear RNAs (9). proteins were predicted using the Database for Annotation, Through high-throughput sequencing, three types of circRNAs Visualization and Integrated Discovery. Gene Ontology have been identified: Exonic circRNAs (7), circular intronic enrichment analysis suggested that the proteins associated RNAs (ciRNAs) (9), and retained-intron circular RNAs or with CDR1as were mainly regulated in the cytoplasm as exon-intron circRNAs (elciRNAs) (10). -
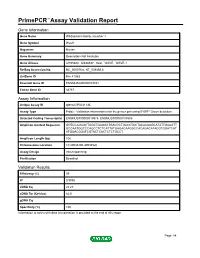
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name WAS protein family, member 1 Gene Symbol Wasf1 Organism Mouse Gene Summary Description Not Available Gene Aliases AI195380, AI838537, Scar, WAVE, WAVE-1 RefSeq Accession No. NC_000076.6, NT_039492.8 UniGene ID Mm.41353 Ensembl Gene ID ENSMUSG00000019831 Entrez Gene ID 83767 Assay Information Unique Assay ID qMmuCIP0031135 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSMUST00000019975, ENSMUST00000105509 Amplicon Context Sequence GGTCCAGAGCTGGCTGAGGATGACGCTGACCTCCTACACAAGCATATTGAAGTT GCCAATGGCCCAGCCTCTCATTATGAGACAAGGCCACAGACATACGTGGATCAT ATGGACGGATCGTACTCACTCTCTGCCT Amplicon Length (bp) 106 Chromosome Location 10:40933200-40934526 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 99 R2 0.9998 cDNA Cq 22.23 cDNA Tm (Celsius) 82.5 gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Wasf1, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

(SCD) Among Indian Patients Using Gene Expression Data Analysis
www.bioinformation.net Hypothesis Volume 14(7) Identification of therapeutic targets for inflammation in sickle cell disease (SCD) among Indian patients using gene expression data analysis Ipsita Das1, Hrishikesh Mishra2, Prafulla K. Khodiar1, Pradeep K. Patra1* 1Pt. J.N.M. Medical College, Raipur, India; 2Sickle Cell Institute Chhattisgarh, Raipur, India; Pradeep K Patra – E-mail: [email protected]; Phone: 91-771-2890012; *Corresponding author Received July 24, 2018; Revised July 29, 2018; Accepted July 29, 2018; Published July 31, 2018 doi: 10.6026/97320630014408 Abstract: Sickle cell disease (SCD) is life-threatening hemoglobinopathy prevalent in India, Sub-Saharan Africa and Middle East. Inflammation plays a pivotal role in disease process and involves intricate interaction among leukocytes, platelets, sickle erythrocytes and vascular endothelium. Available disease modifying therapies are hydroxyl-urea and blood transfusion. Therefore, it is of interest to develop improved pharmacological agents for SCD. We report up-regulated genes in steady state and vaso-occlusive crisis using analysis of gene expression data obtained by microarray experiment for SCD as potential targets. The association of these targets with inflammation in pathway analysis is also documented. Keywords: Sickle cell disease, vaso-occlusive crisis, inflammation, gene expression, pathophysiology, drug targets. Background: vascular endothelium. Leukocytosis and activation of neutrophils Sickle cell disease (SCD) is a life threatening hemoglobin disorder and monocytes further increases vascular inflammation and affecting about 5% of world population and is prevalent in India endothelial damage and plays as a trigger for VOC [3]. In and other parts of the world including Sub-Saharan Africa and addition to vaso-occlusion, inflammation plays a pivotal role in Middle East.