Transcription and Chromatin Organization of a Housekeeping Gene Cluster Containing an Integrated B-Globin Locus Control Region
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Distinctions Between Pediatric and Adult Mature B-Cell Non-Hodgkin Lymphomas Identified Through Genomic Profiling
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Public Health Resources Public Health Resources 4-19-2012 Molecular distinctions between pediatric and adult mature B-cell non-Hodgkin lymphomas identified through genomic profiling Karen Deffenbacher University of Nebraska Medical Center, Omaha Javeed Iqbal University of Nebraska Medical Center, Omaha Warren Sanger University of Nebraska Medical Center, Omaha Yulei Shen University of Nebraska Medical Center, Omaha Cynthia Lachel University of Nebraska Medical Center, Omaha See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/publichealthresources Part of the Public Health Commons Deffenbacher, Karen; Iqbal, Javeed; Sanger, Warren; Shen, Yulei; Lachel, Cynthia; Liu, Zhongfeng; Liu, Yanyan; Lim, Megan; Perkins, Sherrie; Fu, Kai; Smith, Lynette; Lynch, James; Staudt, Louis; Rimsza, Lisa M.; Jaffe, Elaine; Rosenwald, Andreas; Ott, German; Delabie, Jan; Campo, Elias; Gascoyne, Randy; Cairo, Mitchell; Weisenburger, Dennis; Greiner, Timothy; Gross, Thomas; and Chan, Wing, "Molecular distinctions between pediatric and adult mature B-cell non-Hodgkin lymphomas identified through genomic profiling" (2012). Public Health Resources. 145. https://digitalcommons.unl.edu/publichealthresources/145 This Article is brought to you for free and open access by the Public Health Resources at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Public Health Resources by an authorized administrator -

(12) Patent Application Publication (10) Pub. No.: US 2012/0322864 A1 Rossi Et Al
US 2012O322864A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0322864 A1 Rossi et al. (43) Pub. Date: Dec. 20, 2012 (54) SUSTAINED POLYPEPTIDE EXPRESSION (60) Provisional application No. 61/325,003, filed on Apr. FROM SYNTHETIC, MODIFIED RNAS AND 16, 2010, provisional application No. 61/387,220, USES THEREOF filed on Sep. 28, 2010. Publication Classification (75) Inventors: Derrick Rossi, Roslindale, MA (US); (51) Int. Cl. Luigi Warren, Cambridge, MA (US) CI2N5/07 (2010.01) A6IR 48/00 (2006.01) (73) Assignee: IMMUNE DISEASE INSTITUTE, C7H 2L/02 (2006.01) INC., Boston, MA (US) (52) U.S. Cl. ...................... 514/44 R; 536/23.5; 435/377 (57) ABSTRACT (21) Appl. No.: 13/590,880 Described herein are synthetic, modified RNAs for changing the phenotype of a cell. Such as expressing a polypeptide or altering the developmental potential. Accordingly, provided (22) Filed: Aug. 21, 2012 herein are compositions, methods, and kits comprising Syn thetic, modified RNAs for changing the phenotype of a cellor Related U.S. Application Data cells. These methods, compositions, and kits comprising Syn thetic, modified RNAs can be used either to express a desired (63) Continuation of application No. 13/088,009, filed on protein in a cell or tissue, or to change the differentiated Apr. 15, 2011. phenotype of a cell to that of another, desired cell type. Patent Application Publication Dec. 20, 2012 Sheet 1 of 45 US 2012/0322864 A1 s se xxx S s s nnnn s e sit. occa S. s Ra w s e s S. iiii is is iiiiSE SEE SS Patent Application Publication Dec. -

Comparative Analysis of the Ubiquitin-Proteasome System in Homo Sapiens and Saccharomyces Cerevisiae
Comparative Analysis of the Ubiquitin-proteasome system in Homo sapiens and Saccharomyces cerevisiae Inaugural-Dissertation zur Erlangung des Doktorgrades der Mathematisch-Naturwissenschaftlichen Fakultät der Universität zu Köln vorgelegt von Hartmut Scheel aus Rheinbach Köln, 2005 Berichterstatter: Prof. Dr. R. Jürgen Dohmen Prof. Dr. Thomas Langer Dr. Kay Hofmann Tag der mündlichen Prüfung: 18.07.2005 Zusammenfassung I Zusammenfassung Das Ubiquitin-Proteasom System (UPS) stellt den wichtigsten Abbauweg für intrazelluläre Proteine in eukaryotischen Zellen dar. Das abzubauende Protein wird zunächst über eine Enzym-Kaskade mit einer kovalent gebundenen Ubiquitinkette markiert. Anschließend wird das konjugierte Substrat vom Proteasom erkannt und proteolytisch gespalten. Ubiquitin besitzt eine Reihe von Homologen, die ebenfalls posttranslational an Proteine gekoppelt werden können, wie z.B. SUMO und NEDD8. Die hierbei verwendeten Aktivierungs- und Konjugations-Kaskaden sind vollständig analog zu der des Ubiquitin- Systems. Es ist charakteristisch für das UPS, daß sich die Vielzahl der daran beteiligten Proteine aus nur wenigen Proteinfamilien rekrutiert, die durch gemeinsame, funktionale Homologiedomänen gekennzeichnet sind. Einige dieser funktionalen Domänen sind auch in den Modifikations-Systemen der Ubiquitin-Homologen zu finden, jedoch verfügen diese Systeme zusätzlich über spezifische Domänentypen. Homologiedomänen lassen sich als mathematische Modelle in Form von Domänen- deskriptoren (Profile) beschreiben. Diese Deskriptoren können wiederum dazu verwendet werden, mit Hilfe geeigneter Verfahren eine gegebene Proteinsequenz auf das Vorliegen von entsprechenden Homologiedomänen zu untersuchen. Da die im UPS involvierten Homologie- domänen fast ausschließlich auf dieses System und seine Analoga beschränkt sind, können domänen-spezifische Profile zur Katalogisierung der UPS-relevanten Proteine einer Spezies verwendet werden. Auf dieser Basis können dann die entsprechenden UPS-Repertoires verschiedener Spezies miteinander verglichen werden. -

DISCOVERY and CHARACTERIZATION of LONG NON-CODING Rnas in PROSTATE CANCER by John R. Prensner a Dissertation Submitted in Parti
DISCOVERY AND CHARACTERIZATION OF LONG NON-CODING RNAs IN PROSTATE CANCER by John R. Prensner A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Molecular and Cellular Pathology) in the University of Michigan 2012 Doctoral Committee: Professor Arul M. Chinnaiyan, Chair Professor David Beer Professor Kathleen Cho Professor David Ginsburg Assistant Professor Yali Dou © John R. Prensner All Rights Reserved 2012 ACKNOWLEDGEMENTS This work represents the combined efforts of many talented individuals. First, my mentor, Arul Chinnaiyan, deserves much credit for guiding me through this experience and pushing me to produce the best science that I could. My thesis committee—Kathy Cho, David Beer, David Ginsburg, and Yali Dou—was an integral part of my development and I am truly grateful for their participation in my education. I am also indebted to the many collaborators who have contributed to this work. Foremost, Matthew Iyer has been responsible for much of my progress and has been an invaluable colleague. Saravana M. Dhanasekaran mentored me and taught me much of what I know. Wei Chen has spent countless hours working on these projects. I have also been fortunate to collaborate with Felix Feng, Qi Cao, Alejandro Balbin, Sameek Roychowdhury, Brendan Veeneman, Lalit Patel, Anirban Sahu, Chad Brenner, Irfan Asangani, and Xuhong Cao who have helped to make this thesis possible. I would like to extend my appreciation to my wonderful support network at MCTP and the MSTP office: Karen Giles, Christine Betts, Dianna Banka, Ron Koenig, Ellen Elkin, and Hilkka Ketola, who have been immensely supportive and helpful during my training. -

Global Patterns of Changes in the Gene Expression Associated with Genesis of Cancer a Dissertation Submitted in Partial Fulfillm
Global Patterns Of Changes In The Gene Expression Associated With Genesis Of Cancer A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy at George Mason University By Ganiraju Manyam Master of Science IIIT-Hyderabad, 2004 Bachelor of Engineering Bharatiar University, 2002 Director: Dr. Ancha Baranova, Associate Professor Department of Molecular & Microbiology Fall Semester 2009 George Mason University Fairfax, VA Copyright: 2009 Ganiraju Manyam All Rights Reserved ii DEDICATION To my parents Pattabhi Ramanna and Veera Venkata Satyavathi who introduced me to the joy of learning. To friends, family and colleagues who have contributed in work, thought, and support to this project. iii ACKNOWLEDGEMENTS I would like to thank my advisor, Dr. Ancha Baranova, whose tolerance, patience, guidance and encouragement helped me throughout the study. This dissertation would not have been possible without her ever ending support. She is very sincere and generous with her knowledge, availability, compassion, wisdom and feedback. I would also like to thank Dr. Vikas Chandhoke for funding my research generously during my doctoral study at George Mason University. Special thanks go to Dr. Patrick Gillevet, Dr. Alessandro Giuliani, Dr. Maria Stepanova who devoted their time to provide me with their valuable contributions and guidance to formulate this project. Thanks to the faculty of Molecular and Micro Biology (MMB) department, Dr. Jim Willett and Dr. Monique Vanhoek in embedding valuable thoughts to this dissertation by being in my dissertation committee. I would also like to thank the present and previous doctoral program directors, Dr. Daniel Cox and Dr. Geraldine Grant, for facilitating, allowing, and encouraging me to work in this project. -
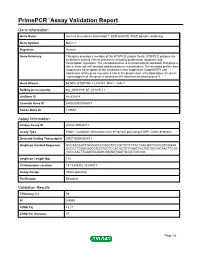
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name nucleus accumbens associated 1, BEN and BTB (POZ) domain containing Gene Symbol NACC1 Organism Human Gene Summary This gene encodes a member of the BTB/POZ protein family. BTB/POZ proteins are involved in several cellular processes including proliferation apoptosis and transcription regulation. The encoded protein is a transcriptional repressor that plays a role in stem cell self-renewal and pluripotency maintenance. The encoded protein also suppresses transcription of the candidate tumor suppressor Gadd45GIP1 and expression of this gene may play a role in the progression of multiple types of cancer. A pseudogene of this gene is located on the short arm of chromosome 9. Gene Aliases BEND8, BTBD14B, FLJ37383, NAC-1, NAC1 RefSeq Accession No. NC_000019.9, NT_011295.11 UniGene ID Hs.531614 Ensembl Gene ID ENSG00000160877 Entrez Gene ID 112939 Assay Information Unique Assay ID qHsaCIP0028717 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000292431 Amplicon Context Sequence GCCAACAGCTGCGGCACCGGCATCCGCTCTTCTACCAACGATCCCCGTCGGAA GCCCCTGGACAGCCGCGTGCTCCACGCTGTCAAGTACTACTGCCAGAACTTCGC CCCCAACTTCAAGGAGAGCGAGATGAATGCCATCGCGG Amplicon Length (bp) 115 Chromosome Location 19:13248302-13249017 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9995 cDNA Cq 19.17 cDNA Tm (Celsius) 87 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 27.88 Specificity (%) 100 Information -

Nac1 (NACC1) (NM 052876) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC208794 Nac1 (NACC1) (NM_052876) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: Nac1 (NACC1) (NM_052876) Human Tagged ORF Clone Tag: Myc-DDK Symbol: NACC1 Synonyms: BEND8; BTBD14B; BTBD30; NAC-1; NAC1; NECFM Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 4 Nac1 (NACC1) (NM_052876) Human Tagged ORF Clone – RC208794 ORF Nucleotide >RC208794 representing NM_052876 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGCCCAGACACTGCAGATGGAGATCCCGAACTTCGGCAACAGCATCCTGGAGTGCCTCAATGAACAGC GGCTGCAGGGCCTGTACTGTGACGTGTCAGTGGTGGTCAAGGGCCATGCCTTCAAGGCCCACCGGGCCGT GCTTGCTGCCAGCAGCTCCTACTTCCGGGACCTGTTCAACAACAGCCGCAGCGCCGTGGTGGAGCTGCCG GCGGCTGTGCAGCCCCAGTCTTTCCAGCAGATCCTCAGCTTCTGCTACACGGGCCGGCTGAGCATGAACG TGGGCGACCAGTTCCTGCTCATGTACACGGCTGGCTTCCTGCAGATCCAGGAGATCATGGAGAAGGGCAC CGAGTTCTTCCTCAAGGTGAGCTCCCCGAGCTGCGACTCCCAGGGCCTGCATGCGGAGGAGGCCCCATCG TCGGAGCCCCAGAGCCCCGTGGCGCAGACATCGGGCTGGCCAGCCTGTAGCACCCCGCTGCCCCTCGTGT CGCGGGTGAAGACGGAGCAGCAGGAGTCGGACTCCGTGCAGTGCATGCCCGTGGCCAAGCGGCTGTGGGA CAGTGGCCAGAAGGAGGCTGGGGGCGGCGGCAATGGCAGCCGCAAGATGGCCAAGTTCTCCACGCCGGAC -

NAC1 Antibody (Human Preferred) A
Revision 1 C 0 2 - t NAC1 Antibody (Human Preferred) a e r o t S Orders: 877-616-CELL (2355) [email protected] Support: 877-678-TECH (8324) 0 2 Web: [email protected] 4 www.cellsignal.com 4 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source: UniProt ID: Entrez-Gene Id: WB H Endogenous 62 Rabbit Q96RE7 112939 Product Usage Information 7. Jinawath, N. et al. (2009) Oncogene 28, 1941-8. 8. Kim, J. et al. (2008) Cell 132, 1049-61. Application Dilution Western Blotting 1:1000 Storage Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA and 50% glycerol. Store at –20°C. Do not aliquot the antibody. Specificity / Sensitivity NAC1 Antibody (Human Preferred) detects endogenous levels of total NAC1 protein. Species Reactivity: Human Species predicted to react based on 100% sequence homology: Monkey Source / Purification Polyclonal antibodies are produced by immunizing animals with a synthetic peptide corresponding to an amino acid sequence at the carboxyl terminus of human NAC1. Antibodies are purified by protein A and peptide affinity chromatography. Background NAC1 or nuclear accumbens-1 is a nuclear factor that belongs to the POZ/BTB (Pox virus and zinc finger/bric-a-brac tramtrack broad complex) domain family. Also known as BTBD14B, it was originally identified in a unique neuronal forebrain structure responsible for reward motivation and addictive behaviors (1,2). NAC1 recruits HDAC3 and HDAC4 to transcriptionally repress gene expression in neuronal cells (3) and specifically co-represses other POZ/BTB proteins in the central nervous system (4). -

Individual Functions and Substrate Specificities of Importin Alpha
From the Max-Delbrück-Center for Molecular Medicine Berlin Head of the research group: Prof. Dr. M. Bader “Individual functions and substrate specificities of importin α subtypes” Dissertation for Fulfillment of Requirements for the Doctoral Degree of the University of Lübeck from the Department of Natural Sciences Submitted by Stefanie Hügel from Hennigsdorf Lübeck 2013 First referee: Prof. Dr. M. Bader Second referee: Prof. Dr. N. Tautz Date of oral examination: 21.02.13 Approved for printing. Lübeck, 22.02.2013 2 1 TABLE OF CONTENTS 1 TABLE OF CONTENTS _______________________________________________________ 3 2 ABSTRACT _______________________________________________________________ 6 3 ZUSAMMENFASSUNG ______________________________________________________ 7 4 INTRODUCTION ___________________________________________________________ 9 4.1 Nucleocytoplasmic protein transport ____________________________________________ 9 4.1.1 The nuclear pore complex – gateway to the nucleus ______________________________________ 10 4.1.1 Transport factors __________________________________________________________________ 11 4.1.2 Nothing happens without RanGTP _____________________________________________________ 12 4.2 The classical nuclear protein import pathway ____________________________________ 13 4.2.1 Structure and function of importin α ___________________________________________________ 13 4.2.2 Importin α:β mediated nuclear protein import___________________________________________ 15 4.2.3 The importin α protein family ________________________________________________________ -

Thesis Corrected-Formatted
Evolutionary, structural and functional features of cellular signalling networks Benjamin Lang Homerton College University of Cambridge Dissertation submitted for the degree of Doctor of Philosophy September 2012 Advisor: Dr. M. Madan Babu Division of Structural Studies Medical Research Council Laboratory of Molecular Biology Declaration of originality I hereby declare that this dissertation is the result of my own work, and neither includes work submitted for the award of a previous degree nor work done in collaboration, except where specifically indicated in the text. I also declare that the text presented does not exceed a length of 60,000 words, excluding appendices, bibliography and figures. Benjamin Lang Cambridge, 27th September, 2012 Evolutionary, structural and functional features of cellular signalling networks Benjamin Lang Summary The post-translational modification of proteins is a fundamental means of biological information processing, with important functions in development, homeostasis and disease. Post-translational modifications (PTMs) can dynamically diversify the proteome in response to intracellular and extracellular signals. Since thousands of modified residues as well as entirely new modification types have recently been discovered in proteins, elucidating their biological functions and identifying the protein components of these PTM systems is a fundamental problem. Chapter 1 gives an overview of the types and known biological functions of different PTMs, as well as experimental methods used to detect them. Intrinsic disorder in proteins is introduced as a structural feature which may influence local evolutionary rates. Several examples of complex PTM signalling systems are then described. Chapter 2 presents a study of the evolution of modified amino acids in human proteins. -

Systematic Analysis, Comparison, and Integration of Disease Based Human
Zhang et al. BMC Medical Genomics 2010, 3:1 http://www.biomedcentral.com/1755-8794/3/1 RESEARCH ARTICLE Open Access Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information Yonqing Zhang1†, Supriyo De1†, John R Garner1, Kirstin Smith1, S Alex Wang2, Kevin G Becker1* Abstract Background: The genetic contributions to human common disorders and mouse genetic models of disease are complex and often overlapping. In common human diseases, unlike classical Mendelian disorders, genetic factors generally have small effect sizes, are multifactorial, and are highly pleiotropic. Likewise, mouse genetic models of disease often have pleiotropic and overlapping phenotypes. Moreover, phenotypic descriptions in the literature in both human and mouse are often poorly characterized and difficult to compare directly. Methods: In this report, human genetic association results from the literature are summarized with regard to replication, disease phenotype, and gene specific results; and organized in the context of a systematic disease ontology. Similarly summarized mouse genetic disease models are organized within the Mammalian Phenotype ontology. Human and mouse disease and phenotype based gene sets are identified. These disease gene sets are then compared individually and in large groups through dendrogram analysis and hierarchical clustering analysis. Results: Human disease and mouse phenotype gene sets are shown to group into disease and phenotypically relevant groups at both a coarse and fine level based on gene sharing. Conclusion: This analysis provides a systematic and global perspective on the genetics of common human disease as compared to itself and in the context of mouse genetic models of disease. -

Gene Trap Cassettes for Random and Targeted
(19) & (11) EP 1 815 001 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/90 (2006.01) A01K 67/027 (2006.01) 02.02.2011 Bulletin 2011/05 C12N 5/10 (2006.01) (21) Application number: 05813398.4 (86) International application number: PCT/EP2005/056282 (22) Date of filing: 28.11.2005 (87) International publication number: WO 2006/056617 (01.06.2006 Gazette 2006/22) (54) GENE TRAP CASSETTES FOR RANDOM AND TARGE TED CONDITIONAL GENE INACTIVATION GEN-FALLEN-KASSETTE FÜR EINE GERICHTETE UND UNGERICHTETE KONDITIONELLE GENINAKTIVIERUNG CASSETTES DE PIEGE A GENES POUR L’INACTIVATION GENETIQUE CONDITIONNELLE ALEATOIRE ET CIBLEE (84) Designated Contracting States: • FLOSS, Thomas AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 85406 Oberappersdorf (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • HANSEN, Jens SK TR 85551 Kirchheim (DE) (30) Priority: 26.11.2004 EP 04028194 (74) Representative: von Kreisler Selting Werner 18.04.2005 EP 05103092 Deichmannhaus am Dom Bahnhofsvorplatz 1 (43) Date of publication of application: 50667 Köln (DE) 08.08.2007 Bulletin 2007/32 (56) References cited: (73) Proprietors: WO-A-02/40685 WO-A-02/088353 • FrankGen Biotechnologie AG 61476 Kronberg/Taunus (DE) • SCHNUETGEN FRANK ET AL: "A directional • Helmholtz Zentrum München Deutsches strategy for monitoring Cre-mediated Forschungszentrum für Gesundheit recombination at the cellular level in the mouse." und Umwelt (GmbH) NATURE BIOTECHNOLOGY, vol. 21, no. 5, May 85764 Neuherberg (DE) 2003 (2003-05), pages 562-565, XP001206846 •MPGMax- Planck-Gesellschaft zur Förderung der ISSN: 1087-0156 Wissenschaften e.V.