International Phd Student Symposium and Career Fair for Life Sciences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New Editor on Journal of Cell Science Michael Way (Editor-In-Chief)
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2019) 132, jcs229740. doi:10.1242/jcs.229740 EDITORIAL New Editor on Journal of Cell Science Michael Way (Editor-in-Chief) As someone who has worked on things related to the actin cytoskeleton my whole research career, the nucleus was not something I paid much attention to. Yes, there were scattered historical reports of actin in the nucleus long before I started my PhD, but no one believed actin was really there of course – it was all an artefact of fixation, you know. Nuclear actin was taboo and no one talked about it at the meetings I went to as a student and postdoc. How wrong we were – today nuclear actin is alive and kicking, although there are definitely more questions than answers concerning what it is actually doing there. We now appreciate that the nucleus contains a wide assortment of proteins associated with the cytoplasmic actin cytoskeleton including myosin motors and actin nucleators such as the Arp2/3 complex. In addition, it should not be forgotten that many chromatin-associated complexes including SWI/SNF and INO80/ SWR also contain multiple actin-related proteins, as well as actin itself. It strikes me that maybe we should all be paying more attention to the nucleus and not just because it contains my favourite proteins! Maybe that’s why, in recent years, we’ve been seeing more submissions to JCS that are focused on different aspects of the nucleus and that traditionally appeared in journals with ‘molecular’ in their titles. -

Cooperative and Antagonistic Contributions of Two Heterochromatin Proteins to Transcriptional Regulation of the Drosophila Sex Determination Decision
Cooperative and Antagonistic Contributions of Two Heterochromatin Proteins to Transcriptional Regulation of the Drosophila Sex Determination Decision Hui Li1, Janel Rodriguez2, Youngdong Yoo1, Momin Mohammed Shareef1, RamaKrishna Badugu1, Jamila I. Horabin2.*, Rebecca Kellum1.* 1 Department of Biology, University of Kentucky, Lexington, Kentucky, United States of America, 2 Department of Biomedical Sciences, Florida State University, Tallahassee, Florida, United States of America Abstract Eukaryotic nuclei contain regions of differentially staining chromatin (heterochromatin), which remain condensed throughout the cell cycle and are largely transcriptionally silent. RNAi knockdown of the highly conserved heterochromatin protein HP1 in Drosophila was previously shown to preferentially reduce male viability. Here we report a similar phenotype for the telomeric partner of HP1, HOAP, and roles for both proteins in regulating the Drosophila sex determination pathway. Specifically, these proteins regulate the critical decision in this pathway, firing of the establishment promoter of the masterswitch gene, Sex-lethal (Sxl). Female-specific activation of this promoter, SxlPe, is essential to females, as it provides SXL protein to initiate the productive female-specific splicing of later Sxl transcripts, which are transcribed from the maintenance promoter (SxlPm) in both sexes. HOAP mutants show inappropriate SxlPe firing in males and the concomitant inappropriate splicing of SxlPm-derived transcripts, while females show premature firing of SxlPe. HP1 mutants, by contrast, display SxlPm splicing defects in both sexes. Chromatin immunoprecipitation assays show both proteins are associated with SxlPe sequences. In embryos from HP1 mutant mothers and Sxl mutant fathers, female viability and RNA polymerase II recruitment to SxlPe are severely compromised. Our genetic and biochemical assays indicate a repressing activity for HOAP and both activating and repressing roles for HP1 at SxlPe. -

The PX Domain Protein Interaction Network in Yeast
The PX domain protein interaction network in yeast Zur Erlangung des akademischen Grades eines DOKTORS DER NATURWISSENSCHAFTEN (Dr. rer. nat.) der Fakultät für Chemie und Biowissenschaften der Universität Karlsruhe (TH) vorgelegte DISSERTATION von Dipl. Biol. Carolina S. Müller aus Buenos Aires Dekan: Prof. Dr. Manfred Kappes Referent: Dr. Nils Johnsson Korreferent: HD. Dr. Adam Bertl Tag der mündlichen Prüfung: 17.02.2005 I dedicate this work to my Parents and Alex TABLE OF CONTENTS Table of contents Introduction 1 Yeast as a model organism in proteome analysis 1 Protein-protein interactions 2 Protein Domains in Yeast 3 Classification of protein interaction domains 3 Phosphoinositides 5 Function 5 Structure 5 Biochemistry 6 Localization 7 Lipid Binding Domains 8 The PX domain 10 Function of PX domain containing proteins 10 PX domain structure and PI binding affinities 10 Yeast PX domain containing proteins 13 PX domain and protein-protein interactions 13 Lipid binding domains and protein-protein interactions 14 The PX-only proteins Grd19p and Ypt35p and their phenotypes 15 Aim of my PhD work 16 Project outline 16 Searching for interacting partners 16 Confirmation of obtained interactions via a 16 second independent method Mapping the interacting region 16 The Two-Hybrid System 17 Definition 17 Basic Principle of the classical Yeast-Two Hybrid System 17 Peptide Synthesis 18 SPOT synthesis technique 18 Analysis of protein- peptide contact sites based on SPOT synthesis 19 TABLE OF CONTENTS Experimental procedures 21 Yeast two-hybrid assay -

Evolution of Gene Dosage on the Z-Chromosome of Schistosome
RESEARCH ARTICLE Evolution of gene dosage on the Z-chromosome of schistosome parasites Marion A L Picard1, Celine Cosseau2, Sabrina Ferre´ 3, Thomas Quack4, Christoph G Grevelding4, Yohann Coute´ 3, Beatriz Vicoso1* 1Institute of Science and Technology Austria, Klosterneuburg, Austria; 2University of Perpignan Via Domitia, IHPE UMR 5244, CNRS, IFREMER, University Montpellier, Perpignan, France; 3Universite´ Grenoble Alpes, CEA, Inserm, BIG-BGE, Grenoble, France; 4Institute for Parasitology, Biomedical Research Center Seltersberg, Justus- Liebig-University, Giessen, Germany Abstract XY systems usually show chromosome-wide compensation of X-linked genes, while in many ZW systems, compensation is restricted to a minority of dosage-sensitive genes. Why such differences arose is still unclear. Here, we combine comparative genomics, transcriptomics and proteomics to obtain a complete overview of the evolution of gene dosage on the Z-chromosome of Schistosoma parasites. We compare the Z-chromosome gene content of African (Schistosoma mansoni and S. haematobium) and Asian (S. japonicum) schistosomes and describe lineage-specific evolutionary strata. We use these to assess gene expression evolution following sex-linkage. The resulting patterns suggest a reduction in expression of Z-linked genes in females, combined with upregulation of the Z in both sexes, in line with the first step of Ohno’s classic model of dosage compensation evolution. Quantitative proteomics suggest that post-transcriptional mechanisms do not play a major role in balancing the expression of Z-linked genes. DOI: https://doi.org/10.7554/eLife.35684.001 *For correspondence: [email protected] Introduction In species with separate sexes, genetic sex determination is often present in the form of differenti- Competing interests: The ated sex chromosomes (Bachtrog et al., 2014). -

Max Planck Researchers Reveal How the DNA Is Made Accessible for Gene Transcription
Public relations Marcus Rockoff Tel: 0761 - 5108 - 368 [email protected] Press release Freiburg i.Br., Who keeps an eye on the house keeper? March 4, 2019 Max Planck researchers reveal how the DNA is made accessible for page 1 of 3 gene transcription It is essential for our cells that the genes on the chromosomes are expressed in the cor- rect dose in each cell type. This also applies to the so-called housekeeping genes, which are necessary for the maintenance of the basic functions of each cell. Researchers at the Max Planck Institute for Immunobiology and Epigenetics now have succeeded in show- ing how transcription is started, especially with household genes, and how the required level of gene dose can be maintained. The study appeared in the journal Genes and Development and uses the fruit fly Drosophila as an example to show how a highly con- served protein complex called NSL positions the nucleosomes in such a way that it is possible for the cell to start reading the genes and control the level of gene expression. In our bodies, there are more than 200 different cell types. Cells take on their identities by switching on distinct sets of genes. But specific genes have to be transcribed in every cell. Scientists refer to them as ‘housekeeping genes’ because their are required for supporting basic cellular function such as metabolism. However, as every organ and cell type is functionally unique, they need a different dose of each active gene to meet their individual requirements. Moreover, a deregulation of these processes can lead to severe health problems or cancer. -

Chemical Agent and Antibodies B-Raf Inhibitor RAF265
Supplemental Materials and Methods: Chemical agent and antibodies B-Raf inhibitor RAF265 [5-(2-(5-(trifluromethyl)-1H-imidazol-2-yl)pyridin-4-yloxy)-N-(4-trifluoromethyl)phenyl-1-methyl-1H-benzp{D, }imidazol-2- amine] was kindly provided by Novartis Pharma AG and dissolved in solvent ethanol:propylene glycol:2.5% tween-80 (percentage 6:23:71) for oral delivery to mice by gavage. Antibodies to phospho-ERK1/2 Thr202/Tyr204(4370), phosphoMEK1/2(2338 and 9121)), phospho-cyclin D1(3300), cyclin D1 (2978), PLK1 (4513) BIM (2933), BAX (2772), BCL2 (2876) were from Cell Signaling Technology. Additional antibodies for phospho-ERK1,2 detection for western blot were from Promega (V803A), and Santa Cruz (E-Y, SC7383). Total ERK antibody for western blot analysis was K-23 from Santa Cruz (SC-94). Ki67 antibody (ab833) was from ABCAM, Mcl1 antibody (559027) was from BD Biosciences, Factor VIII antibody was from Dako (A082), CD31 antibody was from Dianova, (DIA310), and Cot antibody was from Santa Cruz Biotechnology (sc-373677). For the cyclin D1 second antibody staining was with an Alexa Fluor 568 donkey anti-rabbit IgG (Invitrogen, A10042) (1:200 dilution). The pMEK1 fluorescence was developed using the Alexa Fluor 488 chicken anti-rabbit IgG second antibody (1:200 dilution).TUNEL staining kits were from Promega (G2350). Mouse Implant Studies: Biopsy tissues were delivered to research laboratory in ice-cold Dulbecco's Modified Eagle Medium (DMEM) buffer solution. As the tissue mass available from each biopsy was limited, we first passaged the biopsy tissue in Balb/c nu/Foxn1 athymic nude mice (6-8 weeks of age and weighing 22-25g, purchased from Harlan Sprague Dawley, USA) to increase the volume of tumor for further implantation. -

Spatial and Temporal Aspects of Signalling 6 1
r r r Cell Signalling Biology Michael J. Berridge Module 6 Spatial and Temporal Aspects of Signalling 6 1 Module 6 Spatial and Temporal Aspects of Signalling Synopsis The function and efficiency of cell signalling pathways are very dependent on their organization both in space and time. With regard to spatial organization, signalling components are highly organized with respect to their cellular location and how they transmit information from one region of the cell to another. This spatial organization of signalling pathways depends on the molecular interactions that occur between signalling components that use signal transduction domains to construct signalling pathways. Very often, the components responsible for information transfer mechanisms are held in place by being attached to scaffolding proteins to form macromolecular signalling complexes. Sometimes these macromolecular complexes can be organized further by being localized to specific regions of the cell, as found in lipid rafts and caveolae or in the T-tubule regions of skeletal and cardiac cells. Another feature of the spatial aspects concerns the local Another important temporal aspect is timing and signal and global aspects of signalling. The spatial organization of integration, which relates to the way in which functional signalling molecules mentioned above can lead to highly interactions between signalling pathways are determined localized signalling events, but when the signalling mo- by both the order and the timing of their presentations. lecules are more evenly distributed, signals can spread The organization of signalling systems in both time and more globally throughout the cell. In addition, signals space greatly enhances both their efficiency and versatility. -

Modular Phosphoinositide-Binding Domains – Their Role in Signalling and Membrane Trafficking Peter J
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector R882 Review Modular phosphoinositide-binding domains – their role in signalling and membrane trafficking Peter J. Cullen*, Gyles E. Cozier*, George Banting† and Harry Mellor† The membrane phospholipid phosphatidylinositol is the Introduction precursor of a family of lipid second-messengers, The membrane phospholipid phosphatidylinositol is the known as phosphoinositides, which differ in the precursor for a family of lipid second-messengers, known phosphorylation status of their inositol group. A major collectively as phosphoinositides, that differ solely in the advance in understanding phosphoinositide signalling phosphorylation status of their inositol head group has been the identification of a number of highly (Figure 1) [1,2]. Phosphoinositides are ideally suited to conserved modular protein domains whose function function as spatially restricted membrane second messen- appears to be to bind various phosphoinositides. Such gers because: the synthesis and turnover of phosphoinosi- ‘cut and paste’ modules are found in a diverse array of tides from the relatively abundant phosphatidylinositol multidomain proteins and recruit their host protein to membrane precursor can be rapid and highly concentrated specific regions in cells via interactions with within discrete membrane micro domains; the ratio of phosphoinositides. Here, with particular reference to phosphoinositide to binding partner can be relatively large, -
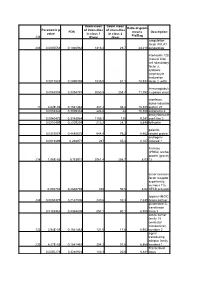
238 Parametric P- Value FDR Geom Mean of Intensities in Class 1 (Plate
Geom mean Geom mean Ratio of geom Parametric p- of intensities of intensities FDR means Description value in class 1 in class 2 Pla/Bag 238 (Plate) (Bag) coagulation factor XIII, A1 206 0,0005558 0,1940962 1313,3 29,7 44,219 polypeptide interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation 0,0017883 0,2490398 1535,5 81,1 18,933 factor 2, p40) Immunoglobuli 0,0063804 0,3394787 3050,5 258,7 11,792 n epsilon chain interferon, alpha-inducible 11 4,62E-05 0,1541482 347,2 33,4 10,395 protein 27 0,0035346 0,3039332 226,8 22 10,309 astrotactin 2 deoxyribonucle 0,0040473 0,3148984 1155,1 125 9,241 ase I-like 3 0,0010495 0,2205294 213,2 24,1 8,846 follistatin galectin- 0,0315078 0,4885675 644,8 76,2 8,462 related protein androgen- 0,0013899 0,238971 287 35,4 8,107 induced 1 A kinase (PRKA) anchor protein (gravin) 236 1,06E-05 0,115911 2061,4 256,7 8,03 12 tumor necrosis factor receptor superfamily, member 11a, 0,002746 0,2846759 405 50,5 8,02 NFKB activator lipoma HMGIC 248 0,0008375 0,2147006 240,6 32,3 7,449 fusion partner glutathione S- transferase 0,0183462 0,4368809 557,1 80,1 6,955 theta 1 solute carrier family 18 (vesicular monoamine), 122 3,94E-05 0,1541482 121,5 17,6 6,903 member 2 signal transducing adaptor family 233 6,27E-05 0,1541482 354,3 51,6 6,866 member 1 Transcribed 0,0050276 0,3240923 140,3 20,6 6,811 locus aldehyde dehydrogenas e 5 family, member A1 (succinate- semialdehyde dehydrogenas 0,0294865 0,4819829 236,6 38,2 6,194 e) guanylate cyclase activator 1A 0,0074099 0,3504479 339,1 55 6,165 -

Research Interests – Dr. Asifa Akhtar
Research Interests – Dr. Asifa Akhtar Every cell in the human body has the same genetic information, collectively known as the genome. In spite of this, your body is composed of highly specialized organs with distinct functions, for example, the heart or brain. At the molecular level, we know that functional biomolecules, RNA and proteins, are responsible for the phenotypic differences between these organs. Since the genome contains instructions for the synthesis of all RNA and pro- teins, this begs the question of how the organism chooses which ones are produced by a particular cell at a given time. Understanding how genetic information is decoded in our cells is the central question driving research in my lab. A cell’s choice to produce a particular RNA or protein is driven by epigenetic regulation. In simple terms, epigenetics is the study of changes in gene expression, which are not caused by changes in the DNA sequence. If phrased in an analogy, the genome is a cookbook filled with recipes (genes), and the epigenetic regulator is the chef responsible for choosing the recipe and for opening or closing and bookmarking the book at the appropriate page, which either facilitates or prevents the recipe from being read. My lab is interested in the molecular components and mechanisms driving epigenetic regula- tion. More specifically, I am fascinated by epigenetic regulation through histone acetylation and large non-coding RNAs. Histones are proteins that are intimately associated with and regulate access to the genome. Histone acetylation refers to the addition of a chemical label to these histone proteins that acts as a signpost or marker to indicate to the cell which gene should get expressed. -

Distinct Mechanisms Mediate X Chromosome Dosage Compensation in Anopheles and Drosophila
Published Online: 15 July, 2021 | Supp Info: http://doi.org/10.26508/lsa.202000996 Downloaded from life-science-alliance.org on 25 September, 2021 Distinct mechanisms mediate X chromosome dosage compensation in Anopheles and Drosophila Claudia Keller Valsecchi, Eric Marois, M. Basilicata, Plamen Georgiev, and Asifa Akhtar DOI: https://doi.org/10.26508/lsa.20200996 Corresponding author(s): Asifa Akhtar, Max Planck Institute of Immunobiology and Epigenetics and Claudia Keller Valsecchi, Institute of Molecular Biology (IMB), Mainz, Germany Review Timeline: Submission Date: 2020-12-16 Editorial Decision: 2021-02-07 Revision Received: 2021-06-03 Editorial Decision: 2021-06-18 Revision Received: 2021-06-27 Accepted: 2021-06-28 Scientific Editor: Eric Sawey, PhD Transaction Report: (Note: With the exception of the correction of typographical or spelling errors that could be a source of ambiguity, letters and reports are not edited. The original formatting of letters and referee reports may not be reflected in this compilation.) 1st Editorial Decision February 7, 2021 February 7, 2021 Re: Life Science Alliance manuscript #LSA-2020-00996-T Asifa Akhtar Max Planck Institute of Immunobiology and Epigenetics Dear Dr. Akhtar, Thank you for submitting your manuscript entitled "Distinct mechanisms mediate X chromosome dosage compensation in Anopheles and Drosophila" to Life Science Alliance. The manuscript was assessed by expert reviewers, whose comments are appended to this letter. As you will note from the reviewers comments below, both reviewers are overall positive about the manuscript, but have raised some interesting and important points that need to be responded to prior to further consideration of the manuscript. -

Deciphering the BAR Code of Membrane Modulators
Cell. Mol. Life Sci. DOI 10.1007/s00018-017-2478-0 Cellular and Molecular LifeSciences REVIEW Deciphering the BAR code of membrane modulators Ulrich Salzer1 · Julius Kostan2 · Kristina Djinović‑Carugo2,3 Received: 30 September 2016 / Revised: 25 January 2017 / Accepted: 27 January 2017 © The Author(s) 2017. This article is published with open access at Springerlink.com Abstract The BAR domain is the eponymous domain of Keywords N-BAR domain · F-BAR domain · I-BAR the “BAR-domain protein superfamily”, a large and diverse domain · lipid binding · Membrane remodelling · set of mostly multi-domain proteins that play eminent roles Membrane curvature at the membrane cytoskeleton interface. BAR domain homodimers are the functional units that peripherally asso- ciate with lipid membranes and are involved in membrane Introduction sculpting activities. Differences in their intrinsic curvatures and lipid-binding properties account for a large variety in Identification of sequence homology between the N-termi- membrane modulating properties. Membrane activities nal regions of the Bin, the amphiphysin, and the yeast Rvs of BAR domains are further modified and regulated by proteins led to the recognition of a novel protein domain intramolecular or inter-subunit domains, by intermolecu- which was named “BAR domain” as an acronym composed lar protein interactions, and by posttranslational modifica- of the first letters of these proteins [1]. This domain was tions. Rather than providing detailed cell biological infor- found in a large set of proteins which were classified as mation on single members of this superfamily, this review BAR domain proteins, later also termed N-BAR domain focuses on biochemical, biophysical, and structural aspects proteins, because several members of this protein family and on recent findings that paradigmatically promote our have an amphipathic helix at the N-terminus of the BAR understanding of processes driven and modulated by BAR domain [2].