Rai, H.S., Reeves, P.A., Peakall, R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Book of Abstracts.Pdf
1 List of presenters A A., Hudson 329 Anil Kumar, Nadesa 189 Panicker A., Kingman 329 Arnautova, Elena 150 Abeli, Thomas 168 Aronson, James 197, 326 Abu Taleb, Tariq 215 ARSLA N, Kadir 363 351Abunnasr, 288 Arvanitis, Pantelis 114 Yaser Agnello, Gaia 268 Aspetakis, Ioannis 114 Aguilar, Rudy 105 Astafieff, Katia 80, 207 Ait Babahmad, 351 Avancini, Ricardo 320 Rachid Al Issaey , 235 Awas, Tesfaye 354, 176 Ghudaina Albrecht , Matthew 326 Ay, Nurhan 78 Allan, Eric 222 Aydınkal, Rasim 31 Murat Allenstein, Pamela 38 Ayenew, Ashenafi 337 Amat De León 233 Azevedo, Carine 204 Arce, Elena An, Miao 286 B B., Von Arx 365 Bétrisey, Sébastien 113 Bang, Miin 160 Birkinshaw, Chris 326 Barblishvili, Tinatin 336 Bizard, Léa 168 Barham, Ellie 179 Bjureke, Kristina 186 Barker, Katharine 220 Blackmore, 325 Stephen Barreiro, Graciela 287 Blanchflower, Paul 94 Barreiro, Graciela 139 Boillat, Cyril 119, 279 Barteau, Benjamin 131 Bonnet, François 67 Bar-Yoseph, Adi 230 Boom, Brian 262, 141 Bauters, Kenneth 118 Boratyński, Adam 113 Bavcon, Jože 111, 110 Bouman, Roderick 15 Beck, Sarah 217 Bouteleau, Serge 287, 139 Beech, Emily 128 Bray, Laurent 350 Beech, Emily 135 Breman, Elinor 168, 170, 280 Bellefroid, Elke 166, 118, 165 Brockington, 342 Samuel Bellet Serrano, 233, 259 Brockington, 341 María Samuel Berg, Christian 168 Burkart, Michael 81 6th Global Botanic Gardens Congress, 26-30 June 2017, Geneva, Switzerland 2 C C., Sousa 329 Chen, Xiaoya 261 Cable, Stuart 312 Cheng, Hyo Cheng 160 Cabral-Oliveira, 204 Cho, YC 49 Joana Callicrate, Taylor 105 Choi, Go Eun 202 Calonje, Michael 105 Christe, Camille 113 Cao, Zhikun 270 Clark, John 105, 251 Carta, Angelino 170 Coddington, 220 Carta Jonathan Caruso, Emily 351 Cole, Chris 24 Casimiro, Pedro 244 Cook, Alexandra 212 Casino, Ana 276, 277, 318 Coombes, Allen 147 Castro, Sílvia 204 Corlett, Richard 86 Catoni, Rosangela 335 Corona Callejas , 274 Norma Edith Cavender, Nicole 84, 139 Correia, Filipe 204 Ceron Carpio , 274 Costa, João 244 Amparo B. -
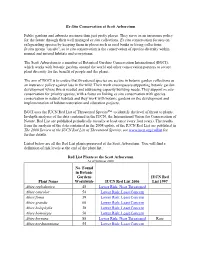
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details. -

Rare Or Threatened Vascular Plant Species of Wollemi National Park, Central Eastern New South Wales
Rare or threatened vascular plant species of Wollemi National Park, central eastern New South Wales. Stephen A.J. Bell Eastcoast Flora Survey PO Box 216 Kotara Fair, NSW 2289, AUSTRALIA Abstract: Wollemi National Park (c. 32o 20’– 33o 30’S, 150o– 151oE), approximately 100 km north-west of Sydney, conserves over 500 000 ha of the Triassic sandstone environments of the Central Coast and Tablelands of New South Wales, and occupies approximately 25% of the Sydney Basin biogeographical region. 94 taxa of conservation signiicance have been recorded and Wollemi is recognised as an important reservoir of rare and uncommon plant taxa, conserving more than 20% of all listed threatened species for the Central Coast, Central Tablelands and Central Western Slopes botanical divisions. For a land area occupying only 0.05% of these divisions, Wollemi is of paramount importance in regional conservation. Surveys within Wollemi National Park over the last decade have recorded several new populations of signiicant vascular plant species, including some sizeable range extensions. This paper summarises the current status of all rare or threatened taxa, describes habitat and associated species for many of these and proposes IUCN (2001) codes for all, as well as suggesting revisions to current conservation risk codes for some species. For Wollemi National Park 37 species are currently listed as Endangered (15 species) or Vulnerable (22 species) under the New South Wales Threatened Species Conservation Act 1995. An additional 50 species are currently listed as nationally rare under the Briggs and Leigh (1996) classiication, or have been suggested as such by various workers. Seven species are awaiting further taxonomic investigation, including Eucalyptus sp. -

Kew Science Publications for the Academic Year 2017–18
KEW SCIENCE PUBLICATIONS FOR THE ACADEMIC YEAR 2017–18 FOR THE ACADEMIC Kew Science Publications kew.org For the academic year 2017–18 ¥ Z i 9E ' ' . -,i,c-"'.'f'l] Foreword Kew’s mission is to be a global resource in We present these publications under the four plant and fungal knowledge. Kew currently has key questions set out in Kew’s Science Strategy over 300 scientists undertaking collection- 2015–2020: based research and collaborating with more than 400 organisations in over 100 countries What plants and fungi occur to deliver this mission. The knowledge obtained 1 on Earth and how is this from this research is disseminated in a number diversity distributed? p2 of different ways from annual reports (e.g. stateoftheworldsplants.org) and web-based What drivers and processes portals (e.g. plantsoftheworldonline.org) to 2 underpin global plant and academic papers. fungal diversity? p32 In the academic year 2017-2018, Kew scientists, in collaboration with numerous What plant and fungal diversity is national and international research partners, 3 under threat and what needs to be published 358 papers in international peer conserved to provide resilience reviewed journals and books. Here we bring to global change? p54 together the abstracts of some of these papers. Due to space constraints we have Which plants and fungi contribute to included only those which are led by a Kew 4 important ecosystem services, scientist; a full list of publications, however, can sustainable livelihoods and natural be found at kew.org/publications capital and how do we manage them? p72 * Indicates Kew staff or research associate authors. -

Pdf Environment and Conservation, Hurstville
References and further reading Scientific Committee (2015). Hibbertia sp. Turramurra Final Determination NSW Threatened Species Scientific Committee. Keith, D.A. (2004). Ocean shores to desert dunes: the native Available from www.environment.nsw.gov.au/resources/ vegetation of New South Wales and the A.C.T. Department of threatenedspecies/determinations/FDHibbTurraCR.pdf Environment and Conservation, Hurstville. Threatened Species Scientific Committee (2016). Conservation OEH (2013). The Native Vegetation of the Sydney Metropolitan Advice Hibbertia spanantha Julian’s hibbertia. Department of the Area (Version 2.0). Office of Environment and Heritage, Environment and Energy, Canberrra. Department of Premier and Cabinet, Sydney. Toelken, H.R. and Robinson, A.F. (2015). Notes on Hibbertia OEH (2017). Julian’s Hibbertia-profile. Office of Environment (Dilleniaceae) 11. Hibbertia spanantha, a new species from the and Heritage NSW Government, Sydney. Accessed 6th central coast of New South Wales. Journal of the Adelaide Botanic February 2018 Available from www.environment.nsw.gov.au/ Gardens 29: 11–14 threatenedSpeciesApp/profile.aspx?id=20279 Threatened plant translocation case study: Wollemia nobilis (Wollemi Pine), Araucariaceae HEIDI ZIMMER1*, PATRICK BAKER2, CATHERINE OFFORD3, JESSICA RIGG4, GREG BOURKE5 AND TONY AULD1 1 NSW Office of Environment and Heritage 2 University of Melbourne 3 The Australian Botanic Garden Mount Annan 4 NSW Department of Primary Industries 5 The Blue Mountains Botanic Garden Mount Tomah *Corresponding author: [email protected] The species There are around 200 seedlings and juvenile Wollemi Pines in the wild. It is likely that the creation of canopy The Wollemi Pine is a Critically Endangered conifer that is gaps would increase Wollemi Pine recruitment, as many endemic to a single catchment in Wollemi National Park, rainforest trees. -

Gymnosperms on the EDGE Félix Forest1, Justin Moat 1,2, Elisabeth Baloch1, Neil A
www.nature.com/scientificreports OPEN Gymnosperms on the EDGE Félix Forest1, Justin Moat 1,2, Elisabeth Baloch1, Neil A. Brummitt3, Steve P. Bachman 1,2, Stef Ickert-Bond 4, Peter M. Hollingsworth5, Aaron Liston6, Damon P. Little7, Sarah Mathews8,9, Hardeep Rai10, Catarina Rydin11, Dennis W. Stevenson7, Philip Thomas5 & Sven Buerki3,12 Driven by limited resources and a sense of urgency, the prioritization of species for conservation has Received: 12 May 2017 been a persistent concern in conservation science. Gymnosperms (comprising ginkgo, conifers, cycads, and gnetophytes) are one of the most threatened groups of living organisms, with 40% of the species Accepted: 28 March 2018 at high risk of extinction, about twice as many as the most recent estimates for all plants (i.e. 21.4%). Published: xx xx xxxx This high proportion of species facing extinction highlights the urgent action required to secure their future through an objective prioritization approach. The Evolutionary Distinct and Globally Endangered (EDGE) method rapidly ranks species based on their evolutionary distinctiveness and the extinction risks they face. EDGE is applied to gymnosperms using a phylogenetic tree comprising DNA sequence data for 85% of gymnosperm species (923 out of 1090 species), to which the 167 missing species were added, and IUCN Red List assessments available for 92% of species. The efect of diferent extinction probability transformations and the handling of IUCN data defcient species on the resulting rankings is investigated. Although top entries in our ranking comprise species that were expected to score well (e.g. Wollemia nobilis, Ginkgo biloba), many were unexpected (e.g. -

Wollemi Pine (Wollemia Nobilis) and Its Introduction to Cultivation in Great Britain and Ireland©
432 Combined Proceedings International Plant Propagators’ Society, Volume 58, 2008 Wollemi Pine (Wollemia nobilis) and Its Introduction to Cultivation in Great Britain and Ireland© Mark Taylor Kernock Park Plants, Pillaton, Saltash, Cornwall PL12 6RY Email: [email protected] INTRODUCTION The Wollemi pine (Wollemia nobilis) was discovered within the Wollemi National Park, a virtually untouched wilderness area of 361,000 ha in the Blue Mountains, 200 km north west of Sydney Australia. It was found on 10 Sept. 1994, when a park ranger was exploring the deep canyons in the park. Wollemi is an Australian Ab- original word which means “stop, and look around you.” The ranger, David Noble, knew most of the tree species in the Park and he must have followed this Aboriginal saying when he stumbled across a grove of enormous trees unlike any that he had seen before. When I spoke to him on his visit to the U.K. in April 2006, he said it was a very strange sensation being in that canyon and even then, it evoked feelings and thoughts of the fairly recent film of that time, Jurassic Park. Noble collected a section of foliage to show his colleagues at the New South Wales National Parks and Wildlife Service. It was soon established that the tree belonged to the Araucariaceae, the same family as the monkey puzzle, Norfolk Island pine, and less well known trees such as the hoop, bunya, and kauri pines. Further inves- tigation, including help from the Royal Botanic Gardens, Sydney, revealed that this was, in fact, a completely new genus of tree. -

Wollemia Nobilis – an Update JONATHON JONES, Garden Director at Tregothnan in Cornwall, Describes the First Flowering of This Species in England
TREES Wollemia nobilis – an update JONATHON JONES, Garden Director at Tregothnan in Cornwall, describes the first flowering of this species in England. Wollemia nobilis first hit the headlines in 1994 when David Noble found the grove of them northwest of Sydney, the exact location still secret. Tregothnan had been busy adding to its collection of rarities for and immediately set about adding this Araucariaceae family star as soon as possible. During the late nineties an exciting rush of new material came to Tregothnan from the International Conifer Conservation Programme and it was hoped that this highly respected route would transcend the bureaucracy and allow in the first ‘Pinosaur’, as it was now called. This was not to be. The Australians mounted the finest example of commercial conservation the world had yet seen. The setting up of the Wollemi Pine conservation club and the Wollemi Pine website communicated the benefits of sensitive commercial propagation of the trees and a coordinated release across the world. Tregothnan was conducting a tea tasting at the Royal Botanic Gardens Kew in October 2005 with its own botanical first, tea actually grown in the UK. This had attracted huge international interest with crews from USA and Japan keen to see The Honourable Evelyn Boscawen about his tea and other plants his family had pioneered at Tregothnan since 1335. But what really caught Mr Boscawen’s eye were the posters advertising the forthcoming auction of the Wollemia. 47 When the auction got underway in Sydney Mr Boscawen was a registered telephone bidder and succeeded in purchasing an early lot, a 1.5m tree of great promise, shortly after 6am. -

Wollemia Nobilis (Wollemi Pine)
1 Wollemia nobilis W.G.Jones, K.D.Hill & J.M.Allen (Wollemi Pine) (Araucariaceae) Distribution: Endemic to NSW Current EPBC Act Status: Endangered Current NSW TSC Act Status: Critically Endangered IUCN RED LIST status: Critically Endangered Proposed change for alignment: update EPBC Act to Critically Endangered Conservation Advice: Wollemia nobilis (Wollemi Pine). Summary of Conservation Assessment Wollemia nobilis was found to be eligible for listing as Critically Endangered under Criterion B via B1ab (iii) (v) and B2ab (iii) (v), Criterion C via C2 (a) (ii); The main reasons for the species being eligible for listing in the Critically Endangered category are i) that the species has a very highly restricted geographic range. The area of occupancy (AOO) and extent of occurrence were estimated to be 4 km2. The AOO is based on one 2 x 2 km grid cell, the scale recommended by the IUCN (2016). The species is only known to exist at one location. Continuing decline is estimated in geographic distribution (extent of occurrence and area of occupancy), area and extent and quality of habitat and the number of mature individuals; ii) less than 250 mature plants are known AND 90-100% of individuals are in one population. Assessment against IUCN Red List criteria Criterion A Population Size reduction. Assessment Outcome: criterion not met. Data deficient. Justification: There is insufficient data to estimate if there is a reduction in the size of the Wollemia nobilis population. The species is long lived (>200-400 years) (DEC 2006) but has only been known for just over 20 years. Therefore, the species is not eligible for listing in any category under this criterion and is data deficient. -

Phytochemistry, Chemotaxonomy, and Biological Activities of the Araucariaceae Family—A Review
plants Review Phytochemistry, Chemotaxonomy, and Biological Activities of the Araucariaceae Family—A Review Claudio Frezza 1,* , Alessandro Venditti 2 , Daniela De Vita 1, Chiara Toniolo 1, Marco Franceschin 2, Antonio Ventrone 1, Lamberto Tomassini 1 , Sebastiano Foddai 1, Marcella Guiso 2, Marcello Nicoletti 1, Armandodoriano Bianco 2 and Mauro Serafini 1 1 Dipartimento di Biologia Ambientale, Università di Roma “La Sapienza”, Piazzale Aldo Moro 5, 00185 Rome, Italy; [email protected] (D.D.V.); [email protected] (C.T.); [email protected] (A.V.); [email protected] (L.T.); [email protected] (S.F.); [email protected] (M.N.); mauro.serafi[email protected] (M.S.) 2 Dipartimento di Chimica, Università di Roma “La Sapienza”, Piazzale Aldo Moro 5, 00185 Rome, Italy; [email protected] (A.V.); [email protected] (M.F.); [email protected] (M.G.); [email protected] (A.B.) * Correspondence: [email protected] Received: 22 June 2020; Accepted: 9 July 2020; Published: 14 July 2020 Abstract: In this review article, the phytochemistry of the species belonging to the Araucariaceae family is explored. Among these, in particular, it is given a wide overview on the phytochemical profile of Wollemia genus, for the first time. In addition to this, the ethnopharmacology and the general biological activities associated to the Araucariaceae species are singularly described. Lastly, the chemotaxonomy at the genus and family levels is described and detailed. Keywords: Araucariaceae; phytochemistry; ethnopharmacology; chemotaxonomy; biological activities 1. Introduction Araucariaceae Henkel and W. Hochstetter is a family of coniferous trees, classified under the order Pinales, the class Pinopsoda, the division Pinophyta, and the Clade Tracheophytes [1]. -

Structure and Abnormalities in Cones of the Wollemi Pine (Wollemia Nobilis)
Erschienen in: Kew Bulletin ; 74 (2019), 1. - 3. https://dx.doi.org/10.1007/s12225-018-9789-7 Structure and abnormalities in cones of the Wollemi pine (Wollemia nobilis ) Veit M. Dörken 1 & Paula J. Rudall 2 Summary. Reproductive structures of both genders of Wollemia nobilis were investigated, including both wild-type and teratological cones. Typically, both pollen cones and seed cones in this species are terminal on ଏrst order branches. At maturity, wild-type pollen cones are pendulous and cylindrical; wild-type seed cones are broad and ellipsoidal in shape. The teratological structures consisted of a basal region that resembled a typical fertile seed cone, and an apical proliferation that terminated in a well-developed pollen cone, resulting in a ‘bisexual’ unit. The proximal seed cone and the distal pollen cone were separated by a sterile region that represents an elongation of the cone axis. Of a total of 14 anomalous bisexual units investigated, all had the same bauplan. Such an arrangement of basal ovulate and distal staminate reproductive structures in a teratological conifer cone has previously not been reported for Wollemia. This topology is 'inside-out' with respect to most other reported anomalous bisexual conifer cones, which possess proximal staminate and distal ovulate structures. We discuss these spontaneous abnormalities in the broader context of understanding the homologies of seed-plant reproductive structures. The patterning of conifer cones is apparently highly labile, perhaps related to the extended cone axis and relatively long developmental duration. Key Words. Araucariaceae, bisexuality, conifers, evolution, morphology, teratology. Introduction 2013). In Agathis and Wollemia, the seeds are winged, Wollemi pine (Wollemia nobilis W. -

Wollemi Pine (Wollemia Nobilis) Recovery Plan
Recovery Plan Number 1 Wollemi Pine (Wollemia nobilis) Recovery Plan September 1998 © NSW National Parks and Wildlife Service, 1998. This work is copyright. Apart from any use as permitted under the Copyright Act 1968, no part may be reproduced without prior written permission from NPWS. NSW National Parks and Wildlife Service 43 Bridge Street (PO Box 1967) HURSTVILLE NSW 2220 Tel: 02 95856444 www.npws.nsw.gov.au For further information contact Threatened Species Unit, Sydney Zone. NSW National Parks and Wildlife Service P.O. Box 1967 HURSTVILLE NSW 2220 Tel 02 9585 6978 Cover illustration: Wollemi Pine (Wollemia nobilis) female cone. Cover illustrator: David Mackay This plan should be cited as following NPWS (1998) ‘Wollemi Pine Recovery Plan’, NPWS, Sydney. ISBN: 0731076354 NSW National Parks and Wildlife Service Recovery Planning Program Wollemi Pine (Wollemia nobilis) Recovery Plan Prepared in accordance with the New South Wales Threatened Species Conservation Act 1995 September 1998 Acknowledgments The research into, and management of, the Wollemi Pine has been a joint effort of the NSW National Parks and Wildlife Service (NPWS) and the Royal Botanic Gardens, Sydney (RBG). This Recovery Plan has been the combined effort of many people who have contributed to the survey and research on the species. The NPWS would like to thank the following people; David Noble (NPWS) who discovered the species; Sharon Nash and Julie Ravallion (NPWS) who prepared this plan; Wyn Jones (NPWS) who contributed to the field work for the ecological monitoring