Genome-Wide Identification, Evolution and Expression of the Complete
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

Supplementary File 2A Revised
Supplementary file 2A. Differentially expressed genes in aldosteronomas compared to all other samples, ranked according to statistical significance. Missing values were not allowed in aldosteronomas, but to a maximum of five in the other samples. Acc UGCluster Name Symbol log Fold Change P - Value Adj. P-Value B R99527 Hs.8162 Hypothetical protein MGC39372 MGC39372 2,17 6,3E-09 5,1E-05 10,2 AA398335 Hs.10414 Kelch domain containing 8A KLHDC8A 2,26 1,2E-08 5,1E-05 9,56 AA441933 Hs.519075 Leiomodin 1 (smooth muscle) LMOD1 2,33 1,3E-08 5,1E-05 9,54 AA630120 Hs.78781 Vascular endothelial growth factor B VEGFB 1,24 1,1E-07 2,9E-04 7,59 R07846 Data not found 3,71 1,2E-07 2,9E-04 7,49 W92795 Hs.434386 Hypothetical protein LOC201229 LOC201229 1,55 2,0E-07 4,0E-04 7,03 AA454564 Hs.323396 Family with sequence similarity 54, member B FAM54B 1,25 3,0E-07 5,2E-04 6,65 AA775249 Hs.513633 G protein-coupled receptor 56 GPR56 -1,63 4,3E-07 6,4E-04 6,33 AA012822 Hs.713814 Oxysterol bining protein OSBP 1,35 5,3E-07 7,1E-04 6,14 R45592 Hs.655271 Regulating synaptic membrane exocytosis 2 RIMS2 2,51 5,9E-07 7,1E-04 6,04 AA282936 Hs.240 M-phase phosphoprotein 1 MPHOSPH -1,40 8,1E-07 8,9E-04 5,74 N34945 Hs.234898 Acetyl-Coenzyme A carboxylase beta ACACB 0,87 9,7E-07 9,8E-04 5,58 R07322 Hs.464137 Acyl-Coenzyme A oxidase 1, palmitoyl ACOX1 0,82 1,3E-06 1,2E-03 5,35 R77144 Hs.488835 Transmembrane protein 120A TMEM120A 1,55 1,7E-06 1,4E-03 5,07 H68542 Hs.420009 Transcribed locus 1,07 1,7E-06 1,4E-03 5,06 AA410184 Hs.696454 PBX/knotted 1 homeobox 2 PKNOX2 1,78 2,0E-06 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
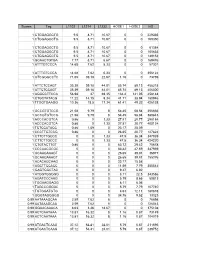
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

RPSA Gene Ribosomal Protein SA
RPSA gene ribosomal protein SA Normal Function The RPSA gene provides instructions for making a protein called ribosomal protein SA, which is one of approximately 80 different ribosomal proteins. These proteins come together to form structures called ribosomes. Ribosomes process the cell's genetic instructions to create proteins. Each ribosome is made up of two parts (subunits) called the large subunit and the small subunit. Ribosomal protein SA is part of the small subunit. The specific roles of each of the ribosomal proteins within the ribosome are not entirely understood. Some ribosomal proteins are involved in the assembly or stability of ribosomes. Others help carry out the ribosome's main function of building new proteins. Research suggests that ribosomal protein SA helps the ribosome control the production of certain proteins, many of which are likely important for development before birth. Health Conditions Related to Genetic Changes Isolated congenital asplenia At least 20 RPSA gene mutations have been identified in individuals with isolated congenital asplenia. People with this condition do not have a spleen but have no other developmental abnormalities. The spleen plays an important role in the immune system. Without this organ, affected individuals are highly susceptible to bacterial infections, which can be life-threatening. RPSA gene mutations are thought to reduce the amount of functional ribosomal protein SA. A shortage of the normal protein likely impairs the assembly of ribosomes, but the specific effects of the mutations -

Ribosomal Proteins Expression and Phylogeny in Alpaca (Lama Pacos) Skin*
Advances in Bioscience and Biotechnology, 2012, 3, 231-237 ABB http://dx.doi.org/10.4236/abb.2012.33032 Published Online June 2012 (http://www.SciRP.org/journal/abb/) Ribosomal proteins expression and phylogeny in alpaca * (Lama pacos) skin Junming Bai, Ruiwen Fan, Shanshan Yang, Yuankai Ji, Jianshan Xie, Chang-Sheng Dong# College of Animal Science and Veterinary Medicine, Shanxi Agricultural University, Taigu, China Email: #[email protected] Received 3 February 2012; revised 8 March 2012; accepted 24 April 2012 ABSTRACT folding/chaperone role in facilitating the processing and folding of rRNA during biogenesis and stabilizing the Ribosomal proteins (RP) has been reported as a cen- mature particle during protein synthesis [3]. It has been tral player in the translation system, and are required well established that global regulation of protein synthe- for the growth and maintenance of all cell kinds. RP sis in eukaryotes is mainly achieved by posttranslational genes form a family of homologous proteins that ex- modification (PTM) of translation factors in response to press in the stable pattern and were used for phy- environmental cues [4]. logenetic analysis. Here we constructed a cDNA li- The family of South American camelids with four brary of alpaca skin and 13,800 clones were sequenced. members is recognized as two wild species, the gua- In the cDNA library, RP genes from skin cDNA li- naco (Lama guanicoe) and the vicuna (Vicugna vicugna) brary of alpaca were identified. Then 8 RP genes were and two of domestic species, the alpaca (Lama pacos) selected at random and built the phylogenetic trees and the llama [5]. -

Inflammation Leads to Distinct Populations of Extracellular Vesicles from Microglia Yiyi Yang1* , Antonio Boza-Serrano1, Christopher J
Yang et al. Journal of Neuroinflammation (2018) 15:168 https://doi.org/10.1186/s12974-018-1204-7 RESEARCH Open Access Inflammation leads to distinct populations of extracellular vesicles from microglia Yiyi Yang1* , Antonio Boza-Serrano1, Christopher J. R. Dunning2, Bettina Hjelm Clausen3,4, Kate Lykke Lambertsen3,4,5 and Tomas Deierborg1* Abstract Background: Activated microglia play an essential role in inflammatory responses elicited in the central nervous system (CNS). Microglia-derived extracellular vesicles (EVs) are suggested to be involved in propagation of inflammatory signals and in the modulation of cell-to-cell communication. However, there is a lack of knowledge on the regulation of EVs and how this in turn facilitates the communication between cells in the brain. Here, we characterized microglial EVs under inflammatory conditions and investigated the effects of inflammation on the EV size, quantity, and protein content. Methods: We have utilized western blot, nanoparticle tracking analysis (NTA), and mass spectrometry to characterize EVs and examine the alterations of secreted EVs from a microglial cell line (BV2) following lipopolysaccharide (LPS) and tumor necrosis factor (TNF) inhibitor (etanercept) treatments, or either alone. The inflammatory responses were measured with multiplex cytokine ELISA and western blot. We also subjected TNF knockout mice to experimental stroke (permanent middle cerebral artery occlusion) and validated the effect of TNF inhibition on EV release. Results: Our analysis of EVs originating from activated BV2 microglia revealed a significant increase in the intravesicular levels of TNF and interleukin (IL)-6. We also observed that the number of EVs released was reduced both in vitro and in vivo when inflammation was inhibited via the TNF pathway. -

Inhibition of the MID1 Protein Complex
Matthes et al. Cell Death Discovery (2018) 4:4 DOI 10.1038/s41420-017-0003-8 Cell Death Discovery ARTICLE Open Access Inhibition of the MID1 protein complex: a novel approach targeting APP protein synthesis Frank Matthes1,MoritzM.Hettich1, Judith Schilling1, Diana Flores-Dominguez1, Nelli Blank1, Thomas Wiglenda2, Alexander Buntru2,HannaWolf1, Stephanie Weber1,InaVorberg 1, Alina Dagane2, Gunnar Dittmar2,3,ErichWanker2, Dan Ehninger1 and Sybille Krauss1 Abstract Alzheimer’s disease (AD) is characterized by two neuropathological hallmarks: senile plaques, which are composed of amyloid-β (Aβ) peptides, and neurofibrillary tangles, which are composed of hyperphosphorylated tau protein. Aβ peptides are derived from sequential proteolytic cleavage of the amyloid precursor protein (APP). In this study, we identified a so far unknown mode of regulation of APP protein synthesis involving the MID1 protein complex: MID1 binds to and regulates the translation of APP mRNA. The underlying mode of action of MID1 involves the mTOR pathway. Thus, inhibition of the MID1 complex reduces the APP protein level in cultures of primary neurons. Based on this, we used one compound that we discovered previously to interfere with the MID1 complex, metformin, for in vivo experiments. Indeed, long-term treatment with metformin decreased APP protein expression levels and consequently Aβ in an AD mouse model. Importantly, we have initiated the metformin treatment late in life, at a time-point where mice were in an already progressed state of the disease, and could observe an improved behavioral phenotype. These 1234567890 1234567890 findings together with our previous observation, showing that inhibition of the MID1 complex by metformin also decreases tau phosphorylation, make the MID1 complex a particularly interesting drug target for treating AD. -

RPSA Antibody / 40S Ribosomal Protein SA (R32562)
RPSA Antibody / 40S Ribosomal protein SA (R32562) Catalog No. Formulation Size R32562 0.5mg/ml if reconstituted with 0.2ml sterile DI water 100 ug Bulk quote request Availability 1-3 business days Species Reactivity Human Format Antigen affinity purified Clonality Polyclonal (rabbit origin) Isotype Rabbit IgG Purity Antigen affinity Buffer Lyophilized from 1X PBS with 2.5% BSA and 0.025% sodium azide UniProt P08865 Applications Western blot : 0.5-1ug/ml Immunohistochemistry (FFPE) : 1-2ug/ml Immunofluorescence (FFPE) : 2-4ug/ml Flow cytometry : 1-3ug/million cells Limitations This RPSA antibody is available for research use only. Western blot testing of human 1) HeLa and 2) U-2 OS cell lysate with RPSA antibody at 0.5ug/ml. Routinely observed molecular weight: 37-40 kDa and 67 kDa. Immunofluorescent staining of FFPE human U-2 OS cells with RPSA antibody (green) and DAPI nuclear stain (blue). HIER: boil tissue sections in pH6, 10mM citrate buffer, for 20 min and allow to cool before testing. IHC staining of FFPE human breast cancer with RPSA antibody. HIER: boil tissue sections in pH6, 10mM citrate buffer, for 20 min and allow to cool before testing. Flow cytometry testing of human A431 cells with RPSA antibody at 1ug/million cells (blocked with goat sera); Red=cells alone, Green=isotype control, Blue= RPSA antibody. Flow cytometry testing of human PC-3 cells with RPSA antibody at 1ug/million cells (blocked with goat sera); Red=cells alone, Green=isotype control, Blue= RPSA antibody. Description 40S ribosomal protein SA is a Ribosomal protein that in humans is encoded by the RPSA gene. -

Ribosomopathies: How a Common Root Can Cause a Tree of Pathologies Nadia Danilova1,* and Hanna T
© 2015. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2015) 8, 1013-1026 doi:10.1242/dmm.020529 REVIEW Ribosomopathies: how a common root can cause a tree of pathologies Nadia Danilova1,* and Hanna T. Gazda2,3,4,* ABSTRACT DBA is a rare disease with an incidence of ∼5 cases per million Defects in ribosome biogenesis are associated with a group of diseases live births, but it has attracted substantial attention as a model called the ribosomopathies, of which Diamond-Blackfan anemia (DBA) disease for ribosomopathies, a group of pathologies associated with is the most studied. Ribosomes are composed of ribosomal proteins defects in ribosome biogenesis (Armistead and Triggs-Raine, 2014; (RPs) and ribosomal RNA (rRNA). RPs and multiple other factors are James et al., 2014). Despite this common defect, phenotypes of necessary for the processing of pre-rRNA, the assembly of ribosomal ribosomopathies differ. A common feature among several subunits, their export to the cytoplasm and for the final assembly of ribosomopathies is p53 activation (Danilova et al., 2008b; subunits into a ribosome. Haploinsufficiency of certain RPs causes Elghetany and Alter, 2002; Jones et al., 2008), but the mechanisms DBA, whereas mutations in other factors cause various other involved have not been completely elucidated. A p53-independent ribosomopathies. Despite the general nature of their underlying response to RP deficiency has also been observed (Aspesi et al., 2014; defects, the clinical manifestations of ribosomopathies differ. In DBA, Danilova et al., 2008b; Singh et al., 2014; Torihara et al., 2011). The for example, red blood cell pathology is especially evident. -

Datasheet: VPA00701 Product Details
Datasheet: VPA00701 Description: RABBIT ANTI RIBOSOMAL PROTEIN SA Specificity: RIBOSOMAL PROTEIN SA Format: Purified Product Type: PrecisionAb™ Polyclonal Isotype: Polyclonal IgG Quantity: 100 µl Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Species Cross Reacts with: Mouse Reactivity N.B. Antibody reactivity and working conditions may vary between species. Product Form Purified IgG - liquid Preparation Rabbit polyclonal antibody purified by affinity chromatography Buffer Solution TRIS buffered glycine Preservative 0.01% Thiomersal Stabilisers <50% Glycerol Immunogen Recombinant protein encompassing part of the middle region of human ribosomal protein SA External Database Links UniProt: P08865 Related reagents Entrez Gene: 3921 RPSA Related reagents Synonyms LAMBR, LAMR1 Page 1 of 2 Specificity Rabbit anti Human ribosomal protein SA antibody recognizes the 40S ribosomal protein SA, also known as 37/67 kDa laminin receptor, LBP/p40, colon carcinoma laminin-binding protein, lamR or LBP. Laminins, a family of extracellular matrix glycoproteins, are the major noncollagenous constituent of basement membranes. -

RPSA, a Candidate Gene for Isolated Congenital Asplenia, Is Required for Pre-Rrna Processing and Spleen Formation in Xenopus John N
© 2018. Published by The Company of Biologists Ltd | Development (2018) 145, dev166181. doi:10.1242/dev.166181 RESEARCH REPORT RPSA, a candidate gene for isolated congenital asplenia, is required for pre-rRNA processing and spleen formation in Xenopus John N. Griffin1, Samuel B. Sondalle2, Andrew Robson1, Emily K. Mis1, Gerald Griffin1, Saurabh S. Kulkarni1, Engin Deniz1, Susan J. Baserga2,3,* and Mustafa K. Khokha1,2,* ABSTRACT 2004; Henras et al., 2008; Woolford and Baserga, 2013). Production A growing number of tissue-specific inherited disorders are starts in the nucleolus, with transcription of pre-ribosomal RNAs associated with impaired ribosome production, despite the (pre-rRNA), and continues through a series of carefully regulated universal requirement for ribosome function. Recently, mutations in cleavages and ribonucleoparticle assembly steps to ultimately RPSA, a protein component of the small ribosomal subunit, were produce the large 60S ribonucleoprotein subunit (LSU, an discovered to underlie approximately half of all isolated congenital intricate complex of 46 ribosomal proteins and the 28S, 5.8S and asplenia cases. However, the mechanisms by which mutations in this 5S rRNAs) and the small 40S subunit (SSU, 33 ribosomal proteins ribosome biogenesis factor lead specifically to spleen agenesis and the 18S rRNA) (Armistead and Triggs-Raine, 2014; Henras remain unknown, in part due to the lack of a suitable animal model for et al., 2008; Sondalle and Baserga, 2014). Along with cytoplasmic study. Here we reveal that RPSA is required for normal spleen translation factors, the LSU and SSU form a functional ribosome. development in the frog, Xenopus tropicalis. Depletion of Rpsa in It is not surprising that defects in ribosome production or function early embryonic development disrupts pre-rRNA processing and are detrimental to health.