Identifying DNA Methylation Biomarkers for Non-Endoscopic Detection of Barrett's Esophagus Helen R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mouse Models of Gastric Cancer
Cancers 2013, 5, 92-130; doi:10.3390/cancers5010092 OPEN ACCESS cancers ISSN 2072-6694 www.mdpi.com/journal/cancers Review Mouse Models of Gastric Cancer Yoku Hayakawa 1, James G. Fox 2, Tamas Gonda 1, Daniel L. Worthley 1, Sureshkumar Muthupalani 2 and Timothy C. Wang 1,* 1 Department of Medicine and Irving Cancer Research Center, Columbia University Medical Center, New York, NY 10032, USA 2 Division of Comparative Medicine, MIT, Cambridge, MA 02139, USA * Author to whom correspondence should be addressed; E-Mail: [email protected]. Received: 5 December 2012; in revised form: 8 January 2013 / Accepted: 15 January 2013 / Published: 24 January 2013 Abstract: Animal models have greatly enriched our understanding of the molecular mechanisms of numerous types of cancers. Gastric cancer is one of the most common cancers worldwide, with a poor prognosis and high incidence of drug-resistance. However, most inbred strains of mice have proven resistant to gastric carcinogenesis. To establish useful models which mimic human gastric cancer phenotypes, investigators have utilized animals infected with Helicobacter species and treated with carcinogens. In addition, by exploiting genetic engineering, a variety of transgenic and knockout mouse models of gastric cancer have emerged, such as INS-GAS mice and TFF1 knockout mice. Investigators have used the combination of carcinogens and gene alteration to accelerate gastric cancer development, but rarely do mouse models show an aggressive and metastatic gastric cancer phenotype that could be relevant to preclinical studies, which may require more specific targeting of gastric progenitor cells. Here, we review current gastric carcinogenesis mouse models and provide our future perspectives on this field. -

ORIGINAL ARTICLE Morphologic Changes of Middle Ear Mucosa In
The Mediterranean Journal of Otology ORIGINAL ARTICLE Morphologic changes of middle ear mucosa in chronic otitis media with or without cholesteatoma Sertaç Yetifler, Yusuf H›d›r, M. Salih Deveci Ac›badem Hospital, Bursa, (S. Yetifler), TURKEY, Gulhane Medical School, Dept of ORL & OBJECTIVE: To investigate histopathologic differences between chronic HNS, Etlik-Ankara, (Y. H›d›r), Gulhane Medical School, Dept of otitis media (COM) with cholesteatoma and COM without cholesteatoma. Pathology, Etlik-Ankara, (M.S. MATER‹ALS AND METHODS: This retrospective study is an analysis of 74 Deveci), TURKEY middle ear biopsies from the promontory near the round window taken at Correspondent Author: first operation for COM performed. Thirty ears had COM with Yusuf Hidir, MD cholesteatoma. The other 44 ears had COM without cholesteatoma. Gulhane Medical School Materials were stained by Hematoxylin-eosin and Toluidine blue. Density Dept of ORL & HNS of gland and secretory cell, epithelial thickness, number of ciliated cell, 06018 Etlik, Ankara, Turkey infiltration and migration of chronic inflammatory cells (lymphocyte and Tel: +90 312 304 5731 plasma cell) and grade of vascular dilatation and proliferation between the Fax: +90 312 304 5700 patients with or without cholesteatoma were compared. The analysis of E-mail: [email protected] quantitative parameters was performed using Pearson χ2 test. Submitted: 16 December 2007 RESULTS: Infiltration and migration of lymphocyte and plasma cells, and Revised: 08 June 2008 grade of vascular dilatation and proliferation were significantly greater in Accepted: 15 July 2008 ears without cholesteatoma than those with cholesteatoma. Mediterr J Otol 2008; 4: 102-108 CONCLUS‹ONS: These findings indicate that distinct physiopathologic mechanisms may play role in development of COM in terms of presence of cholesteatoma. -

Essentials of Pap Smear and Breast Cytology
Essentials of Pap Smear and Breast Cytology Brenda Smith Gia-Khanh Nguyen 2012 Essentials of Pap Smear and Breast Cytology Brenda Smith, BSc, RT, CT (ASCP) Clinical Instructor Department of Pathology & Laboratory Medicine University of British Columbia Vancouver, British Columbia, Canada And Gia-Khanh Nguyen, MD, FRCPC Professor Emeritus Department of Laboratory Medicine & Pathology University of Alberta Edmonton, Alberta, Canada All rights reserved. Legally deposited at Library and Archives Canada. ISBN: 978-0- 9780929-7-9. 2 Table of contents Preface 4 Acknowledgements and Related material by the same author 5 Abbreviations and Remarks 6 Chapter 1. Pap smear: An overview 7 Chapter 2. Pap smear: Normal uterus and vagina 18 Chapter 3. Pap smear: Negative for intraepithelial lesion or malignancy: Infections and nonneoplastic findings 28 Chapter 4. Pap smear: Squamous cell abnormalities 51 Chapter 5. Pap smear: Glandular cell abnormalities 69 Chapter 6. Pap smear: Other malignant tumors 90 Chapter 7. Anal Pap smear: Anal-rectal cytology 98 Chapter 8. Breast cytology: An overview 102 Chapter 9. Nonneoplastic breast lesions 106 Chapter10. Breast neoplasms 116 The authors 146 3 Preface This monograph “Essentials of Pap Smear and Breast Cytology” is prepared at the request of a large number of students in cytology who wish to have a small and concise book with numerous illustrations for easy reference during their laboratory training. Most information and illustrations in this book are extracted from the authors’ monograph entitled “Essentials of Gynecologic Cytology”, and they are rearranged according to The Bethesda System-2001. This book should be used in conjunction with the above-mentioned book on gynecologic cytology. -

Combined Intestinal and Squamous Metaplasia of the Renal Pelvis
J Clin Pathol: first published as 10.1136/jcp.22.2.187 on 1 March 1969. Downloaded from J. clin. Path. (1969), 22, 187-191 Combined intestinal and squamous metaplasia of the renal pelvis R. SALM From the Royal Cornwall Hospital (Treliske), Truro, Cornwall SYNOPSIS A case of combined squamous and intestinal metaplasia of the renal pelvis, associated with hydronephrosis, chronic pyelonephritis, and lithiasis, is reported. The changes are considered to be due primarily to mechanical irritation of the pelvic epithelium. The epithelium of the urinary passages occasionally CALCULUS This measured 5 x 4-5 x 3*5 cm and undergoes metaplastic changes. This is seen not weighed 65 g (Fig. 2). Chemical analysis proved it to infrequently in the bladder, but only rarely in the consist mainly of calcium oxalate. ureter. pelvis and Cystic and glandular transfor- MICROSCOPICAL EXAMINATION (67/2319) Sections of the mation of the urinary transitional cell epithelium four blocks cut from the renal parenchyma all show the occurs more frequently than squamous transforma- features of a marked chronic pyelonephritis. There is a tion. In a few instances a combination of both moderate loss of substance, and below the pelvic lining squamous and glandular metaplasia has been ob- there are zones of dense infiltration with lymphocytes copyright. served in the renal pelvis, of which the following and plasma cells, occasionally extending downwards to case is a further example. involve the patchily thickened pelvic smooth muscle coat. The entire central velvety area of the pelvis was cut in CASE REPORT 11 blocks. There are many areas of erosion and chronic inflammation. -

Transurethral Photoselective Vaporization: a Safe and Effective Treatment for Glandular Cystitis
African Journal of Pharmacy and Pharmacology Vol. 6(25), pp. 1870-1873, 8 July, 2012 Available online at http://www.academicjournals.org/AJPP DOI: 10.5897/AJPP12.358 ISSN 1996-0816 ©2012 Academic Journals Full Length Research Paper Transurethral photoselective vaporization: A safe and effective treatment for glandular cystitis Wei Tao*, Yuxi Shan, Dongrong Yang and Wei Qian Department of Urology, the Second Affiliated Hospital of Soochow University, China. Accepted 6 June, 2012 To evaluate the clinical efficacy and safety of transurethral photoselective vaporization in the treatment of glandular cystitis, 125 cases with glandular cystitis were treated with transurethral photoselective vaporization from 2004 to 2010. The patients’ hospital records were reviewed to obtain pre-operative, intra-operative and post-operative data. All the patients were diagnosed with glandular cystitis through pathological biopsy before or during their operation. All the patients were also operated on successfully. Mean operating time was 22 min and mean post-operative hospital stay was 4 days. Obturator nerve reflux, vesical perforation or transurethral resection syndrome were not found. After vaporization, every patient had a urethral catheter indwelling for 1 to 5 days without the need for bladder irrigation. All cases were followed up for 6 to 24 months, of which 123/125 (98.4%) cases were cured and 2/125 (1.6%) cases showed recurrence during this follow-up period. Therefore, transurethral photoselective vaporization is a safe and effective strategy for the treatment of glandular cystitis. Key words: Transurethral photoselective vaporization of prostate (PVP), glandular cystitis (GC). INTRODUCTION Glandular cystitis (GC) is a special type of inflammation al., 2006) began to use the green light laser for treatment in the urinary system with the tendency to malignant of benign prostatic hyperplasia in our hospital and change (Lancelin et al., 2000). -

“Glassy” Cells in Barrett's Mucosa
ANTICANCER RESEARCH 29: 4145-4146 (2009) “Glassy” Cells in Barrett’s Mucosa CARLOS A. RUBIO Gastrointestinal and Liver Pathology Research Laboratory, Department of Pathology, Karolinska Institute and University Hospital, Stockholm, Sweden Abstract. Background: In 1991 we detected glandular Recently, while examining esophageal biopsies with cells with “glassy” cytoplasm (GCs) in human gastric columnar-lined epithelium in humans, we noticed the mucosa having intestinal metaplasia (IM). GCs were presence of glands displaying GCs. subsequently found in the gastric mucosa of baboons. The purpose of the present work was to audit the frequency of Materials and Methods: The frequency of GCs (defined as cases with GCs in a cohort of esophageal biopsies in humans, glandular cells with a homogeneously pale, eosinophilic having at histological examination, columnar-lined mucosa. cytoplasm having a ground-glass appearance) was assessed in 403 human esophageal biopsies with columnar-lined Materials and Methods esophagus. GCs may be in one gland or in a group of glands in metaplastic mucosa. Results: Out of the 403 From the files of the Department of Pathology, 403 consecutive esophageal biopsies, 176 had cardiac glands with or biopsies having columnar-lined esophageal mucosa were retrieved. without oxyntic cells (glandular metaplasia type 1 and 2, Definitions. Three phenotypes of glands with columnar-lined respectively) and the remaining 227 cases additionally had mucosa may occur in the esophageal metaplastic mucosa: with goblet cells (that is IM), glandular metaplasia type 3 cardiac or with fundic glands (referred to as glandular mucosa (namely, Barrett’s mucosa). Four out of the 403 cases type 1 and 2, respectively) and with intestinal metaplastic glands (0.99% ) had glands with GCs; all four cases were recorded (referred to as glandular mucosa type 3). -

Pet Bird Oncology
PET BIRD ONCOLOGY Drury Reavill Zoo/Exotic Pathology Service 2825 KOVR Drive West Sacramento, CA 95605 (Modified and up‐dated, and used with permission from AAV Proceedings 2001) Although neoplastic diseases occur with some frequency in the commonly kept pet birds, published information regarding prognosis and therapy of specific neoplasms is limited. Each case report or study that provides this information, helps us to improve the level of care we can offer our companion avian pets. This review will cover some basic information about select tumour types with attempted therapies and will reference the more recent reports in the avian literature. It is not intended to be all encompassing. For more information, there are extensive published reviews of the reported avian tumours (Leach 1992; Reece 1992; Reavill,2004). Table 1 provides a summary of therapies reported in pet birds. A survey of the tumours diagnosed at our service, Zoo Excotic Pathology Service (ZEPS), will be discussed. SELECTED PET BIRD TUMOURS Haemangiomas and Haemangiosarcomas Haemangiomas are benign tumours of vascular endothelium. The malignant version is haemangiosarcoma, also known as malignant haemangioendothelioma or angiosarcoma. Haemangiomas are more commonly reported in budgerigars (Melopsittacus undulatus) than in other birds, and usually occur in the skin or spleen (Leach 1992; Reece 1992). Splenic and cutaneous haemangiomas have also been reported in African grey parrots (Psittacus erithacus) and an Indian ring‐necked parakeet (Psittacula krameri) (Leach, 1992). Of the haemangiomas we’ve seen (n=19), the majority were removed from the skin (feet, inguinal region, cloaca, side of neck, and wing); one was found in the spleen. -
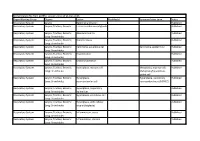
Synonym/Other Term Status Respiratory System Larynx Epithelial Alteration Published Respiratory System Larynx; Trachea; Bronchi Ectasia in Submucosal Glands Published
Nomenclature file from goRENI, version 2.9 as of 21-Dec-2015 Organ Working Group Organs Lesion Modifier(s) Synonym/other term Status Respiratory System Larynx Epithelial alteration Published Respiratory System Larynx; Trachea; Bronchi Ectasia in submucosal glands Published Respiratory System Larynx; Trachea; Bronchi; Adenocarcinoma Published Lung: Bronchioles Respiratory System Larynx; Trachea; Bronchi; Bronchiectasis Published Lung: Bronchioles Respiratory System Larynx; Trachea; Bronchi; Carcinoma, squamous cell Carcinoma, epidermoid Published Lung: Bronchioles Respiratory System Larynx; Trachea; Bronchi; Degeneration Published Lung: Bronchioles Respiratory System Larynx; Trachea; Bronchi; Erosion/ulceration Published Lung: Bronchioles Respiratory System Larynx; Trachea; Bronchi; Hyperplasia, mucous cell Metaplasia, mucous cell; Published Lung: Bronchioles Metaplasia/hyperplasia, goblet cell Respiratory System Larynx; Trachea; Bronchi; Hyperplasia, Hyperplasia, pulmonary Published Lung: Bronchioles neuroendocrine cell neuroendocrine cell (PNEC) Respiratory System Larynx; Trachea; Bronchi; Hyperplasia, respiratory Published Lung: Bronchioles epithelium Respiratory System Larynx; Trachea; Bronchi; Hyperplasia, squamous cell Published Lung: Bronchioles Respiratory System Larynx; Trachea; Bronchi; Hyperplasia, with cellular Published Lung: Bronchioles atypia (dysplasia) Respiratory System Larynx; Trachea; Bronchi; Inflammation, acute Published Lung: Bronchioles Respiratory System Larynx; Trachea; Bronchi; Inflammation, chronic Published -

Histopathology IMPC HIS 003
Histopathology IMPC_HIS_003 Purpose To perform histopathological examination and annotation of the IMPC’s standard list of organs (see IMPReSS Gross Pathology & Tissue Collection) on stained tissue sections oriented on glass slides (see IMPReSS Tissue Embedding & Block Banking). Experimental Design Minimum number of animals : 2M + 2F Age at test: Week 16 Notes Significance Score Not significant . Interpreted by the histopathologist to be a finding attributable to background strain (e.g. low- incidence hydrocephalus, microphthalmia) or incidental to mutant phenotype (e.g. hair- induced glossitis, focal hyperplasia, mild mononuclear cell infiltrate). OR Significant . Interpreted by the histopathologist as a finding not attributable to background strain and not incidental to mutant phenotype. Severity Score Sc Term Definition ore No lesion(s) or abnormalities detectable considering the age and sex of 0 Normal the animal Abnormality visible, involving single or multiple tissue types in a minimal 1 Mild proportion of an organ, likely to have no functional consequence 2 Moder Abnormality visible, involving multiple tissue types in a minority proportion ate of an organ, likely to have no clinical consequence (subclinical) Abnormality clearly visible, involving multiple tissue types in a majority 3 Marked proportion of an organ, likely to have minor clinical manifestation(s) Abnormality clearly visible, involving multiple tissue types in almost all 4 Severe visible area of an organ, likely to have major clinical manifestation(s) Parameters and Metadata Adrenal gland - Descriptor PATO IMPC_HIS_107_001 | v1.0 ontologyParameter Req. Analysis: false Req. Upload: false Is Annotated: false Ontology Options: acute, right, left, subacute, random pattern, multifocal to coalescing, multi-localised, generalized, segmental, distributed, chronic-active, chronic, focally extensive, localized, transmural, bilateral, unilateral, symmetrical, peracute, Adrenal gland - MA term IMPC_HIS_350_001 | v1.0 ontologyParameter Req. -

Cancers of the Upper Gastro-Intestinal Tract: a Review of Somatic Mutation Distributions
*HQH0XWDWLRQVLQ8SSHU*DVWURLQWHVWLQDO&DQFHUV Review Article Cancers of the Upper Gastro-intestinal Tract: A Review of Somatic Mutation Distributions Behnoush Abedi-Ardekani MD13LHUUH+DLQDXW3K'2 Abstract Cancers of the upper gastro-intestinal tract (UGIT) comprise esophageal, esophago-gastric junction, stomach and duodenal cancers. Together, these cancers represent over 1.5 million cases and are the cause of about 1.25 million deaths annually. This group of cancers encompasses diseases with marked disparities in etiology, geographic distribution, histopathological features and frequency. Based on histological origin, squamous cell carcinoma of the esophagus (ESCC), which arises through a dysplasia-carcinoma sequence within the squamous mucosa, is a completely different cancer than junction, stomach and duodenal cancers, which develop within glandular epithelia WKURXJKFDVFDGHVLQYROYLQJLQÀDPPDWLRQPHWDSODVLDG\VSODVLDDQGFDUFLQRPD$WWKHIURQWOLQHEHWZHHQWKHVHWZRKLVWRORJLFDOGRPDLQV cancers of the esophago-gastric junction constitute a mixed group of glandular tumors including distal esophageal adenocarcinomas and cancers arising within the most proximal part of the stomach - the cardia. Most of UGIT cancers are sporadic, although familial susceptibility JHQHVKDYHEHHQLGHQWL¿HGIRUVWRPDFKDQGUDUHFDVHVRI(6&&:HKDYHXVHGWKH&260,&GDWDEDVH http://www.sanger.ac.uk/genetics/ CGP/cosmic/) to identify genes commonly mutated in UGIT cancers. Regardless of etiology and histopathology, three genes are mutated in at least 5% of UGIT cancers: TP53, CDKN2a and PIK3CA. Another three genes, NFE2L2, PTCH1 and NOTCH1, are mutated in ESCC only. Conversely, genes of the RAS family and of the CDH1/APC/CTNNB1 pathway are mutated only in non-squamous cancers, with differ- ences in mutated genes DFFRUGLQJWRWRSRJUDSK\:HUHYLHZWKHSRWHQWLDOIXQFWLRQDOVLJQL¿FDQFHRIthese observations for understanding mechanisms of UGIT carcinogenesis. Key words: Esogastric junction, mutations, upper GI tract cancers Cite this article as: Abedi-Ardekani B, Hainaut P. -

Non-Papillary Urothelial Lesions of the Urinary Bladder: Morphological Classification and Immunohistochemical Markers CONSTANTINA D
in vivo 22 : 493-502 (2008) Review Non-papillary Urothelial Lesions of the Urinary Bladder: Morphological Classification and Immunohistochemical Markers CONSTANTINA D. PETRAKI 1 and CONSTANTINOS P. SFIKAS 2 1Renal Pathology Department, Evangelismos Hospital, Athens; 2Department of Pathology, Helena Venizelou Hospital, Athens, Greece Abstract. In the past quarter century, several nomenclature of view, two basic distinct but occasionally interrelated systems for non-papillary urothelial lesions of the urinary pathways are identified on the basis of the pattern of growth bladder have been proposed. The 1998 World Health of the intraepithelial lesions (papillary and flat), the Organization and International Society of Urological behaviour of these lesions being related to the degree of Pathology (WHO/ISUP) classification recommended a four- architectural and cytological alteration of the urothelium. tier system for the subdivision of flat intraepithelial lesions of “Superficial” lesions (80%) can be divided further into the bladder. This classification was adopted by the WHO in papillary cancer and the much less common “flat” lesions of 1999 and revised in 2004. In 2002, a more detailed high-grade carcinoma in situ (CIS). The remaining 20% of classification of flat urothelial lesions was based on the bladder carcinomas are non-papillary, invasive tumors at the Ancona International Consultation. Each of these lesions time of initial presentation. Reflecting these two distinct was defined with strict morphological criteria to provide clinical -

Proliferative and Nonproliferative Lesions of the Rat and Mouse Hepatobiliary System
Toxicologic Pathology, 38: 5S-81S, 2010 Copyright # 2010 by The Author(s) ISSN: 0192-6233 print / 1533-1601 online DOI: 10.1177/0192623310386499 Proliferative and Nonproliferative Lesions of the Rat and Mouse Hepatobiliary System 1 2* 3 4 5 6 BOB THOOLEN ,ROBERT R. MARONPOT ,TAKANORI HARADA ,ABRAHAM NYSKA ,COLIN ROUSSEAUX ,THOMAS NOLTE , 7 8 9 10 11 12 DAVID E. MALARKEY ,WOLFGANG KAUFMANN ,KARIN KU¨ TTLER ,ULRICH DESCHL ,DAI NAKAE ,RICHARD GREGSON , 13 14 15 16 17 MICHAEL P. VINLOVE ,AMY E. BRIX ,BHANU SINGH ,FIORELLA BELPOGGI , AND JERROLD M. WARD 1Global Pathology Support, The Hague, The Netherlands 2Maronpot Consulting LLC, Raleigh, North Carolina, USA 3The Institute of Environmental Toxicology, Joso-shi, Ibaraki, Japan 4Haharuv 18, Timrat, Israel 5Wakefield QC, Canada 6Boehringer Ingelheim Pharma GmbH & Co., Biberach an der Riss, Germany 7National Toxicology Program, Cellular and Molecular Pathology Branch, Research Triangle Park, North Carolina, USA 8Merck KGaA, Darmstadt, Germany 9BASF Aktiengesellschaft, Ludwigshafen, Germany 10Boehringer Ingelheim Pharma GmbH & Co. KG, Biberach/Riss, Germany 11Tokyo Metropolitan Institute of Public Health, Shinjuku, Tokyo, Japan 12Charles River Laboratories, Pathology Department, Senneville, QC, Canada 13Pathology Associates, Charles River, Frederick, Maryland, USA 14Experimental Pathology Laboratories Inc., Research Triangle Park, North Carolina, USA 15DuPont Haskell Global Centers for Health and Environmental Science, Newark, Delaware, USA 16Ramazzini Institute, Bentivoglio (BO), Italy 17Global VetPathology, Montgomery Village, Maryland, USA *Chairman of the Liver INHAND Committee ABSTRACT The INHAND Project (International Harmonization of Nomenclature and Diagnostic Criteria for Lesions in Rats and Mice) is a joint initiative of the Societies of Toxicologic Pathology from Europe (ESTP), Great Britain (BSTP), Japan (JSTP) and North America (STP) to develop an interna- tionally-accepted nomenclature for proliferative and non-proliferative lesions in laboratory animals.