Clinical Significance of High Expression of Mir-452-5P in Lung Squamous Cell Carcinoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Table S2.Up Or Down Regulated Genes in Tcof1 Knockdown Neuroblastoma N1E-115 Cells Involved in Differentbiological Process Anal
Table S2.Up or down regulated genes in Tcof1 knockdown neuroblastoma N1E-115 cells involved in differentbiological process analysed by DAVID database Pop Pop Fold Term PValue Genes Bonferroni Benjamini FDR Hits Total Enrichment GO:0044257~cellular protein catabolic 2.77E-10 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 537 13588 1.944851 8.64E-07 8.64E-07 5.02E-07 process ISG15, ATG7, PSENEN, LOC100046898, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, ASB8, DCUN1D1, PSMA6, SIAH1A, TRIM32, RNF138, GM12396, RNF20, USP17L5, FBXO11, RAD23B, NEDD8, UBE2V2, RFFL, CDC GO:0051603~proteolysis involved in 4.52E-10 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 534 13588 1.93519 1.41E-06 7.04E-07 8.18E-07 cellular protein catabolic process ISG15, ATG7, PSENEN, LOC100046898, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, ASB8, DCUN1D1, PSMA6, SIAH1A, TRIM32, RNF138, GM12396, RNF20, USP17L5, FBXO11, RAD23B, NEDD8, UBE2V2, RFFL, CDC GO:0044265~cellular macromolecule 6.09E-10 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 609 13588 1.859332 1.90E-06 6.32E-07 1.10E-06 catabolic process ISG15, RBM8A, ATG7, LOC100046898, PSENEN, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, ASB8, DCUN1D1, PSMA6, SIAH1A, TRIM32, RNF138, GM12396, RNF20, XRN2, USP17L5, FBXO11, RAD23B, UBE2V2, NED GO:0030163~protein catabolic process 1.81E-09 MKRN1, PPP2R5C, VPRBP, MYLIP, CDC16, ERLEC1, MKRN2, CUL3, 556 13588 1.87839 5.64E-06 1.41E-06 3.27E-06 ISG15, ATG7, PSENEN, LOC100046898, CDCA3, ANAPC1, ANAPC2, ANAPC5, SOCS3, ENC1, SOCS4, -

Supplementary Table 8. Cpcp PPI Network Details for Significantly Changed Proteins, As Identified in 3.2, Underlying Each of the Five Functional Domains
Supplementary Table 8. cPCP PPI network details for significantly changed proteins, as identified in 3.2, underlying each of the five functional domains. The network nodes represent each significant protein, followed by the list of interactors. Note that identifiers were converted to gene names to facilitate PPI database queries. Functional Domain Node Interactors Development and Park7 Rack1 differentiation Kcnma1 Atp6v1a Ywhae Ywhaz Pgls Hsd3b7 Development and Prdx6 Ncoa3 differentiation Pla2g4a Sufu Ncf2 Gstp1 Grin2b Ywhae Pgls Hsd3b7 Development and Atp1a2 Kcnma1 differentiation Vamp2 Development and Cntn1 Prnp differentiation Ywhaz Clstn1 Dlg4 App Ywhae Ywhab Development and Rac1 Pak1 differentiation Cdc42 Rhoa Dlg4 Ctnnb1 Mapk9 Mapk8 Pik3cb Sod1 Rrad Epb41l2 Nono Ltbp1 Evi5 Rbm39 Aplp2 Smurf2 Grin1 Grin2b Xiap Chn2 Cav1 Cybb Pgls Ywhae Development and Hbb-b1 Atp5b differentiation Hba Kcnma1 Got1 Aldoa Ywhaz Pgls Hsd3b4 Hsd3b7 Ywhae Development and Myh6 Mybpc3 differentiation Prkce Ywhae Development and Amph Capn2 differentiation Ap2a2 Dnm1 Dnm3 Dnm2 Atp6v1a Ywhab Development and Dnm3 Bin1 differentiation Amph Pacsin1 Grb2 Ywhae Bsn Development and Eef2 Ywhaz differentiation Rpgrip1l Atp6v1a Nphp1 Iqcb1 Ezh2 Ywhae Ywhab Pgls Hsd3b7 Hsd3b4 Development and Gnai1 Dlg4 differentiation Development and Gnao1 Dlg4 differentiation Vamp2 App Ywhae Ywhab Development and Psmd3 Rpgrip1l differentiation Psmd4 Hmga2 Development and Thy1 Syp differentiation Atp6v1a App Ywhae Ywhaz Ywhab Hsd3b7 Hsd3b4 Development and Tubb2a Ywhaz differentiation Nphp4 -

Decreased TESK1-Mediated Cofilin 1 Phosphorylation in the Jejunum of IBS-D Patients May Explain Increased Female Predisposition
www.nature.com/scientificreports OPEN Decreased TESK1-mediated coflin 1 phosphorylation in the jejunum of IBS-D patients may explain Received: 22 September 2017 Accepted: 17 January 2018 increased female predisposition to Published: xx xx xxxx epithelial dysfunction Bruno K. Rodiño-Janeiro 1, Cristina Martínez1, Marina Fortea2, Beatriz Lobo1, Marc Pigrau1, Adoración Nieto1, Ana María González-Castro2, Eloísa Salvo-Romero1,2, Danila Guagnozzi2, Cristina Pardo-Camacho2, Cristina Iribarren2, Fernando Azpiroz 1,3, Carmen Alonso-Cotoner1,3, Javier Santos1,3 & Maria Vicario2,3 Disturbed intestinal epithelial barrier and mucosal micro-infammation characterize irritable bowel syndrome (IBS). Despite intensive research demonstrating ovarian hormones modulation of IBS severity, there is still limited knowledge on the mechanisms underlying female predominance in this disorder. Our aim was to identify molecular pathways involved in epithelial barrier dysfunction and female predominance in diarrhea-predominant IBS (IBS-D) patients. Total RNA and protein were obtained from jejunal mucosal biopsies from healthy controls and IBS-D patients meeting the Rome III criteria. IBS severity was recorded based on validated questionnaires. Gene and protein expression profles were obtained and data integrated to explore biological and molecular functions. Results were validated by western blot. Tight junction signaling, mitochondrial dysfunction, regulation of actin-based motility by Rho, and cytoskeleton signaling were diferentially expressed in IBS- D. Decreased TESK1-dependent coflin 1 phosphorylation (pCFL1) was confrmed in IBS-D, which negatively correlated with bowel movements only in female participants. In conclusion, deregulation of cytoskeleton dynamics through TESK1/CFL1 pathway underlies epithelial intestinal dysfunction in the small bowel mucosa of IBS-D, particularly in female patients. -

Protein Expression Changes Induced in a Malignant Melanoma Cell Line
Pisano et al. BMC Cancer (2016) 16:317 DOI 10.1186/s12885-016-2362-6 RESEARCH ARTICLE Open Access Protein expression changes induced in a malignant melanoma cell line by the curcumin analogue compound D6 Marina Pisano1, Antonio Palomba2,3, Alessandro Tanca2, Daniela Pagnozzi2, Sergio Uzzau2, Maria Filippa Addis2, Maria Antonietta Dettori1, Davide Fabbri1, Giuseppe Palmieri1 and Carla Rozzo1* Abstract Background: We have previously demonstrated that the hydroxylated biphenyl compound D6 (3E,3′E) -4,4′-(5,5′,6,6′-tetramethoxy-[1,1′-biphenyl]-3,3′-diyl)bis(but-3-en-2-one), a structural analogue of curcumin, exerts a strong antitumor activity on melanoma cells both in vitro and in vivo. Although the mechanism of action of D6 is yet to be clarified, this compound is thought to inhibit cancer cell growth by arresting the cell cycle in G2/M phase, and to induce apoptosis through the mitochondrial intrinsic pathway. To investigate the changes in protein expression induced by exposure of melanoma cells to D6, a differential proteomic study was carried out on D6-treated and untreated primary melanoma LB24Dagi cells. Methods: Proteins were fractionated by SDS-PAGE and subjected to in gel digestion. The peptide mixtures were analyzed by liquid chromatography coupled with tandem mass spectrometry. Proteins were identified and quantified using database search and spectral counting. Proteomic data were finally uploaded into the Ingenuity Pathway Analysis software to find significantly modulated networks and pathways. Results: Analysis of the differentially expressed protein profiles revealed the activation of a strong cellular stress response, with overexpression of several HSPs and stimulation of ubiquitin-proteasome pathways. -

A Graph-Theoretic Approach to Model Genomic Data and Identify Biological Modules Asscociated with Cancer Outcomes
A Graph-Theoretic Approach to Model Genomic Data and Identify Biological Modules Asscociated with Cancer Outcomes Deanna Petrochilos A dissertation presented in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2013 Reading Committee: Neil Abernethy, Chair John Gennari, Ali Shojaie Program Authorized to Offer Degree: Biomedical Informatics and Health Education UMI Number: 3588836 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. UMI 3588836 Published by ProQuest LLC (2013). Copyright in the Dissertation held by the Author. Microform Edition © ProQuest LLC. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, MI 48106 - 1346 ©Copyright 2013 Deanna Petrochilos University of Washington Abstract Using Graph-Based Methods to Integrate and Analyze Cancer Genomic Data Deanna Petrochilos Chair of the Supervisory Committee: Assistant Professor Neil Abernethy Biomedical Informatics and Health Education Studies of the genetic basis of complex disease present statistical and methodological challenges in the discovery of reliable and high-confidence genes that reveal biological phenomena underlying the etiology of disease or gene signatures prognostic of disease outcomes. This dissertation examines the capacity of graph-theoretical methods to model and analyze genomic information and thus facilitate using prior knowledge to create a more discrete and functionally relevant feature space. -

Xo PANEL DNA GENE LIST
xO PANEL DNA GENE LIST ~1700 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes (at 400 -500x average coverage on tumor) Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 -

The Role of TTP Phosphorylation in the Regulation of Inflammatory Cytokine Production by MK2/3
The Role of TTP Phosphorylation in the Regulation of Inflammatory Cytokine Production by MK2/3 This information is current as Natalia Ronkina, Nelli Shushakova, Christopher Tiedje, of September 28, 2021. Tatiana Yakovleva, Maxim A. X. Tollenaere, Aaron Scott, Tanveer Singh Batth, Jesper Velgaard Olsen, Alexandra Helmke, Simon Holst Bekker-Jensen, Andrew R. Clark, Alexey Kotlyarov and Matthias Gaestel J Immunol published online 16 September 2019 Downloaded from http://www.jimmunol.org/content/early/2019/09/13/jimmun ol.1801221 Supplementary http://www.jimmunol.org/content/suppl/2019/09/13/jimmunol.180122 http://www.jimmunol.org/ Material 1.DCSupplemental Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 28, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2019 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published September 16, 2019, doi:10.4049/jimmunol.1801221 The Journal of Immunology The Role of TTP Phosphorylation in the Regulation of Inflammatory Cytokine Production by MK2/3 Natalia Ronkina,*,1 Nelli Shushakova,†,‡,1 Christopher Tiedje,x Tatiana Yakovleva,* Maxim A. -
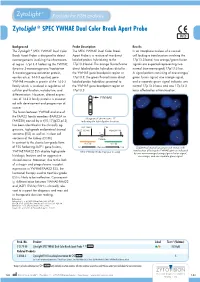
Zytolight ® SPEC YWHAE Dual Color Break Apart Probe
ZytoLight ® SPEC YWHAE Dual Color Break Apart Probe Background Probe Description Results The ZytoLight ® SPEC YWHAE Dual Color The SPEC YWHAE Dual Color Break In an interphase nucleus of a normal Break Apart Probe is designed to detect Apart Probe is a mixture of two direct cell lacking a translocation involving the rearrangements involving the chromosom- labeled probes hybridizing to the 17p13.3 band, two orange/green fusion al region 17p13.3 harboring the YWHAE 17p13.3 band. The orange fluorochrome signals are expected representing two (tyrosine 3-monooxygenase/tryptophan direct labeled probe hybridizes distal to normal (non-rearranged) 17p13.3 loci. 5-monooxygenase activation protein, the YWHAE gene breakpoint region at A signal pattern consisting of one orange/ epsilon a.k.a. 14-3-3 epsilon) gene. 17p13.3, the green fluorochrome direct green fusion signal, one orange signal, YWHAE encodes a protein of the 14-3-3 labeled probe hybridizes proximal to and a separate green signal indicates one family which is involved in regulation of the YWHAE gene breakpoint region at normal 17p13.3 locus and one 17p13.3 cellular proliferation, metabolism, and 17p13.3. locus affected by a translocation. differentiation. However, altered expres- sion of 14-3-3 family proteins is associat- YWHAE ed with development and progression of cancer. The fusion between YWHAE and one of the FAM22 family members (FAM22A or Ideogram of chromosome 17 FAM22B) caused by a t(10;17)(q22;p13) indicating the hybridization locations. has been identified in the clinically ag- RH113855 CRK gressive, high-grade endometrial stromal SHGC-130461 HUMUT6768 sarcoma (ESS) as well as in clear cell 3’ 5’ sarcoma of the kidney (CCSK). -

Proteomic Analysis of Necroptotic Extracellular Vesicles
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.11.037192; this version posted April 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Proteomic analysis of necroptotic extracellular vesicles Inbar Shlomovitz1, Gali Yanovich-Arad2, Ziv Erlich1, Liat Edry-Botzer1, Sefi Zargarian1, Hadar Cohen1, Yifat Ofir-Birin3, Neta Regev-Rudzki3 and Motti Gerlic1 1. Department of Clinical Microbiology and Immunology, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel 2. Department of Human Molecular Genetics and Biochemistry, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel 3. Department of Biomolecular Sciences, Weizmann Institute of Science, Rehovot, Israel Corresponding author Motti Gerlic, Department of Clinical Microbiology and Immunology, Sackler School of Medicine, Tel Aviv University, Tel Aviv 69978, Israel Tel: +972-36409069, E-mail: [email protected] Abstract Necroptosis is a regulated and inflammatory form of cell death. We, and others, have previously reported that necroptotic cells release extracellular vesicles (EVs). We have found that necroptotic EVs are loaded with proteins, including the phosphorylated form of the key necroptosis-executing factor, mixed lineage kinase domain-like kinase (MLKL). However, neither the exact protein composition, nor the impact, of necroptotic EVs have been delineated. To characterize their content, EVs from necroptotic and untreated U937 cells were isolated and analyzed by mass spectrometry-based proteomics. A total of 3337 proteins were identified, sharing a high degree of similarity with exosome proteome databases, and clearly distinguishing necroptotic and control EVs.