Karyotype Variation in Leea L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

List of Hospitals Around the Mumbai Pune Expressway
List of Hospitals around the Mumbai Pune Expressway Name of the Address Contact Details Hospital Pawana Hospital Pawana Hospital Numbers: 02114-221076/221077 [NABH Somatane Phata, Tal-Maval, District Helpline number: +917722049970 accredited] Pune-410506 Emergency Number: +918308817880 [email protected], [email protected] Varad Hospital Varad Hospital, kiwale, Dehu Road, (91)-257-2255561, 2251651 Pune - 412101 JeevanRekha Sr.28 Prabhu Complex, in front of 020 27674777 Hospital Repulic School, Old Mumbai Pune Hwy, (Govt.Hospital) Dehu Road, Maharashtra 412101. Aadhar Multi Old Mumbai - Pune Hwy, Pimpri- 092256 53798 Speciality Chinchwad, Hospital & ICU Maharashtra 412101 (Govt.Hospital) Panacea Hospital Unit 1: 141, Sai Arcade, Mission Unit 1: 9930451695 Compound, Line Ali, Unit2: 91 22 27469999 Panvel, Navi Mumbai-410206 Unit 2: Plot No.105/106, Sector- 8, New Panvel (E) Navi Mumbai-410206 Khalapur Primary Khalapur Primary Health Centre 7768047518 Health Khalapur-410202 [email protected] Centre/Khalapur Khalapur, District Raigarh District Hospital Raigad Hospital And Sau Vandana N.Tasgaonkar Education 02148-221501, 221502 Research Centre Complex, [email protected] Village: Diksal, Post: Koshane, Tal:Karjat, Dist: Raigarh 410201 Opp.Bhivpuri Road Station Maharashtra M G M Hospital 33, Sahitya Mandir Marg, 022- 61526666/Ext-900,901 (NABH Sector 4, Vashi, 022- 61526608/09/10 accredited) Navi Mumbai 27820076 Maharashtra- 400708 Emergency number- 14466 Toll free number- 18002665456 [email protected] Aundh Institute AiMS Hospital & Emergency Research Contact no: +91 (020) 2580 1000 / of Medical Centre 89750 44444 / 020-67400100 Sciences Near AiMS Square, Aundh Pune, Maharashtra=- 411007 Emergency Contact Number: 020 2588 2202 / 2580 1000 Mobile No:+91 89750 44444 Whatsapp 9168657400 Nigidri lokmanya Tilak Road, Sector No. -

Investigation of the Mass Movement in Varand Region, Western Ghat of Ha Maharashtra Using Geospatial Technique
International Journal of Civil Engineering and Technology (IJCIET) Volume 9, Issue 7, July 2018, pp. 20112027, Article ID: IJCIET_09_07_214 Available online at http://iaeme.com/Home/issue/IJCIET?Volume=9&Issue=7 ISSN Print: 0976-6308 and ISSN Online: 0976-6316 © IAEME Publication Scopus Indexed INVESTIGATION OF THE MASS MOVEMENT IN VARANDHA REGION, WESTERN GHAT OF MAHARASHTRA USING GEOSPATIAL TECHNIQUE Dattatraya J. Khamkar PhD Scholar, Civil and Environmental Engineering Department, VJTI, Mumbai-400019, Maharashtra, India Sumedh Y. Mhaske Associate Professor, Civil and Environmental Engineering Department, VJTI, Mumbai-400019, Maharashtra, India ABSTRACT Verandha Ghat Section is connecting shield-plateau region with Konkan Coastal Belt (KCB) through Bhor Ghat area. Therefore, it is considered as a lifeline of Bhor (in Pune district) and Mahad (Raigarh District of Konkan), of western Maharashtra corridor. Geologically, this region belongs to Ambenali and Mahabaleshwar – Poladpur formation. The area shows thick flows of the Deccan Trap basalt of Upper Cretaceous to Eocene age. All the rock flows of this formation, exposed along this highway are in the form of rock cutting along the Pandharpur-Mahad Maharashtra State Highway -70, in the 74 km Ghat section from Bhor (part of Sahyadri Uplands, from Pune district) onwards and before Mahad (part of Konkan Coastal Belt from Raigarh district) of Maharashtra, are highly susceptible for landslide activity. A detailed exploration was carried out on all along the road section started from Bhor city and it extends up to the Varandah village, at the foot hill of Sahyadri, in Konkan region. In the present context, detailed investigations of the mass movement were carried out to prepare the map of highly vulnerable locations, in the study area. -

Reg. No Name in Full Residential Address Gender Contact No. Email Id Remarks 9421864344 022 25401313 / 9869262391 Bhaveshwarikar
Reg. No Name in Full Residential Address Gender Contact No. Email id Remarks 10001 SALPHALE VITTHAL AT POST UMARI (MOTHI) TAL.DIST- Male DEFAULTER SHANKARRAO AKOLA NAME REMOVED 444302 AKOLA MAHARASHTRA 10002 JAGGI RAMANJIT KAUR J.S.JAGGI, GOVIND NAGAR, Male DEFAULTER JASWANT SINGH RAJAPETH, NAME REMOVED AMRAVATI MAHARASHTRA 10003 BAVISKAR DILIP VITHALRAO PLOT NO.2-B, SHIVNAGAR, Male DEFAULTER NR.SHARDA CHOWK, BVS STOP, NAME REMOVED SANGAM TALKIES, NAGPUR MAHARASHTRA 10004 SOMANI VINODKUMAR MAIN ROAD, MANWATH Male 9421864344 RENEWAL UP TO 2018 GOPIKISHAN 431505 PARBHANI Maharashtra 10005 KARMALKAR BHAVESHVARI 11, BHARAT SADAN, 2 ND FLOOR, Female 022 25401313 / bhaveshwarikarmalka@gma NOT RENEW RAVINDRA S.V.ROAD, NAUPADA, THANE 9869262391 il.com (WEST) 400602 THANE Maharashtra 10006 NIRMALKAR DEVENDRA AT- MAREGAON, PO / TA- Male 9423652964 RENEWAL UP TO 2018 VIRUPAKSH MAREGAON, 445303 YAVATMAL Maharashtra 10007 PATIL PREMCHANDRA PATIPURA, WARD NO.18, Male DEFAULTER BHALCHANDRA NAME REMOVED 445001 YAVATMAL MAHARASHTRA 10008 KHAN ALIMKHAN SUJATKHAN AT-PO- LADKHED TA- DARWHA Male 9763175228 NOT RENEW 445208 YAVATMAL Maharashtra 10009 DHANGAWHAL PLINTH HOUSE, 4/A, DHARTI Male 9422288171 RENEWAL UP TO 05/06/2018 SUBHASHKUMAR KHANDU COLONY, NR.G.T.P.STOP, DEOPUR AGRA RD. 424005 DHULE Maharashtra 10010 PATIL SURENDRANATH A/P - PALE KHO. TAL - KALWAN Male 02592 248013 / NOT RENEW DHARMARAJ 9423481207 NASIK Maharashtra 10011 DHANGE PARVEZ ABBAS GREEN ACE RESIDENCY, FLT NO Male 9890207717 RENEWAL UP TO 05/06/2018 402, PLOT NO 73/3, 74/3 SEC- 27, SEAWOODS, -

By Thesis Submitted for the Degree of Vidyavachaspati (Doctor of Philosophy) Faculty for Moral and Social Sciences Department Of
“A STUDY OF AN ECOLOGICAL PATHOLOGICAL AND BIO-CHEMICAL IMPACT OF URBANISATION AND INDUSTRIALISATION ON WATER POLLUTION OF BHIMA RIVER AND ITS TRIBUTARIES PUNE DISTRICTS, MAHARASHTRA, INDIA” BY Dr. PRATAPRAO RAMGHANDRA DIGHAVKAR, I. P. S. THESIS SUBMITTED FOR THE DEGREE OF VIDYAVACHASPATI (DOCTOR OF PHILOSOPHY) FACULTY FOR MORAL AND SOCIAL SCIENCES DEPARTMENT OF SOCIOLOGY TILAK MAHARASHTRA VIDHYAPEETH PUNE JUNE 2016 CERTIFICATE This is to certify that the entire work embodied in this thesis entitled A STUDY OFECOLOGICAL PATHOLOGICAL AND BIOCHEMICAL IMPACT OF URBANISATION AND INDUSTRILISATION ON WATER POLLUTION OF BHIMA RIVER AND Its TRIBUTARIES .PUNE DISTRICT FOR A PERIOD 2013-2015 has been carried out by the candidate DR.PRATAPRAO RAMCHANDRA DIGHAVKAR. I. P. S. under my supervision/guidance in Tilak Maharashtra Vidyapeeth, Pune. Such materials as has been obtained by other sources and has been duly acknowledged in the thesis have not been submitted to any degree or diploma of any University or Institution previously. Date: / / 2016 Place: Pune. Dr.Prataprao Ramchatra Dighavkar, I.P.S. DECLARATION I hereby declare that this dissertation entitled A STUDY OF AN ECOLOGICAL PATHOLOGICAL AND BIO-CHEMICAL IMPACT OF URBANISNTION AND INDUSTRIALISATION ON WATER POLLUTION OF BHIMA RIVER AND Its TRIBUTARIES ,PUNE DISTRICT FOR A PERIOD 2013—2015 is written and submitted by me at the Tilak Maharashtra Vidyapeeth, Pune for the degree of Doctor of Philosophy The present research work is of original nature and the conclusions are base on the data collected by me. To the best of my knowledge this piece of work has not been submitted for the award of any degree or diploma in any University or Institution. -

Drive to Tamhini Ghat During the Monsoon
placesnearpune.com May 27, 2010 Rajaram S Drive to tamhini ghat during the monsoon One such place is the Tamhini Ghat just beyond the Mulshi Dam. Though the roads through this ghat are at best bad, the scenery around would make hen i decided to come back to India (pune) you forget the bumpiness of the ride. A ride through one year ago, i heard zillions of stories this ghat is good even during the summer. But, this Wabout the pollution in Pune, the traffic , the place opens up in the rains. Waterfalls all around, bureacracy at work , all trying to convince me not to greens of the like rarely seen, you have to go there take such a decision. But no one mentioned about to experience this. I drove through Tamhini ghat the abundance of natural beauty around Pune. A late last August on my way to the Hans adventure drive to any such place near Pune is enough to free resort. The other members of my team were in a bus your mind from the stress caused by any of the and i was following them in my Xylo. The tempta- above factors. Eventually, the sight-seeing options tion was too much to stop at every corner and click near to Pune took preference over all other issues a photograph of the panorama. Below are some and helped me make the decision to come to Pune. photos from that drive. print joli Printed with Printed http://www.placesnearpune.com/2010/05/drive-to-tamhini-ghat-during-the-monsoon/ Page 1 placesnearpune.com May 27, 2010 Drive to tamhini ghat during the monsoon How to go to Tamhini Ghat from Pune(Magarpatta): Go to Chandi Chowk. -

41 a Geographical Analysis of Major Tourist Attraction in Pune District, Maharashtra State
I J R S S I S, Vol. V (1), Jan 2017: 41-44 ISSN 2347 – 8268 INTERNATIONAL JOURNAL OF RESEARCHES IN SOCIAL SCIENCES AND INFORMATION STUDIES © VISHWASHANTI MULTIPURPOSE SOCIETY (Global Peace Multipurpose Society) R. No. MH-659/13(N) www.vmsindia.org A GEOGRAPHICAL ANALYSIS OF MAJOR TOURIST ATTRACTION IN PUNE DISTRICT, MAHARASHTRA STATE Amol S. Shinde De pt. of Ge ography, Walchand College of Arts and Science , Solapur (M.S) [email protected] Abstract: The natural resources, coastal lines, waterfalls, hot springs, temples, historical forts, caves, wild-life, hill ranges, scenery and amenable climate are very important resources of tourist attrac-tion.The various facilities available to the domestic and foreign tourists in Pune district. These include natural resources, transportation, infrastructure, hospitality resources and major tourist attractions. For the research work Pune District is selected. This district has at Pune its Satara district at south, Thane and Ahmednagar state at North, Raigad district west, Solapur district east The object of study region is, to highlight the attractive tourist destinations and religious places,Historal,Nature and Cultural Place etc.This study based on primary and secondary data. Tourist attractions in the district as is, natural beauty, caves, temples, forts, mini garden, rock garden, tracking, rock climbing, wild life, festival's fairs, arts, handicrafts, creeks, lakes etc. places. To the stay of tourist, which requires natural resources, infrastructural and transportation facilities, accommodation, food, recreation, sightseeing, shopping and variety of facilities and services for use and enjoyments? The source of tourism depends on all these facilities. Keywords- Pune District, Maharashtra, Tourism, Natural resource Introduction: Pune District, also known as Educational Study Area: Capital of the state of Maharashtra. -
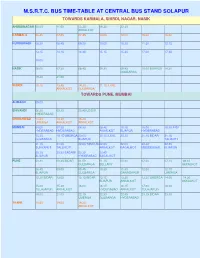
M.S.R.T.C. Bus Time-Table at Central Bus Stand Solapur
M.S.R.T.C. BUS TIME-TABLE AT CENTRAL BUS STAND SOLAPUR TOWARDS KARMALA, SHIRDI, NAGAR, NASIK AHMEDNAGAR 08.00 11.00 13.25 16.30 22.30 AKKALKOT KARMALA 06.45 07.00 07.45 10.00 12.00 15.30 16.00 KURDUWADI 08.30 08.45 09.20 10.00 10.30 11.30 12.15 13.15 14.15 14.45 15.15 15.30 17.00 17.45 18.00 NASIK 06.00 07.30 08.45 09.30 09.45 10.00 BIJAPUR 14.30 GULBARGA 19.30 21.00 SHIRDI 10.15 13.45 14.30 21.15 ILKAL AKKALKOT GULBARGA TOWARDS PUNE, MUMBAI ALIBAGH 09.00 BHIVANDI 06.30 09.30 20.45 UDGIR HYDERABAD CHINCHWAD 13.30 14.30 15.30 UMERGA AKKALKOT AKKALKOT MUMBAI 04.00 07.30 08.30 08.45 10.15 15.00 15.30 INDI HYDERABAD HYDERABAD AKKALKOT BIJAPUR HYDERABAD 15.30 19.15 UMERGA 20.00 20.15 ILKAL 20.30 21.15 BIDAR 21.15 GULBARGA BIJAPUR TALIKOTI 21.15 21.30 22.00 TANDUR 22.00 22.00 22.30 22.45 SURYAPET TALLIKOTI AKKALKOT BAGALKOT MUDDEBIHAL BIJAPUR 23.15 23.30 BADAMI 23.30 23.45 BIJAPUR HYDERABAD BAGALKOT PUNE 00.30 00.45 BIDAR 01.00 01.15 05.30 07.00 07.15 08.15 GULBARGA BELLARY AKKALKOT 08.45 09.00 09.45 10.30 11.30 12.00 12.15 BIJAPUR GULBARGA GANAGAPUR UMERGA 12.30 BIDAR 13.00 13.15 BIDAR 13.15 13.30 13.30 UMERGA 14.00 14.30 BIJAPUR AKKALKOT AKKALKOT 15.00 15.30 16.00 16.15 16.15 17.00 18.00 TULAJAPUR AKKALKOT HYDERABAD AKKALKOT TULAJAPUR 19.00 21.00 22.15 22.30 22.45 23.15 BIDAR 23.30 UMERGA GULBARGA HYDERABAD THANE 10.45 19.00 19.30 AKKALKOT TOWARDS AKKALKOT, GANAGAPUR, GULBARGA AKKALKOT 04.15 05.45 06.00 08.15 09.15 09.15 10.30 10.45 11.00 11.30 11.45 12.15 13.45 14.15 15.30 16.00 16.30 16.45 17.00 GULBARGA 02.00 PUNE 05.15 06.15 07.30 08.15 -

“A Study of Responsible Factors for Instant Price Hike in Properties in Talegaon Dabhade -Pune”
© 2018 JETIR April 2018, Volume 5, Issue 4 www.jetir.org (ISSN-2349-5162) “A STUDY OF RESPONSIBLE FACTORS FOR INSTANT PRICE HIKE IN PROPERTIES IN TALEGAON DABHADE -PUNE” Author -Prof.Dr Rajesh kumar jha Associate professor (Dr. D.Y. Patil Institute of Management and Entrepreneur development –Talegaon, PUNE) ABSTRACT: Real estate has contributed a lot in the reinforcement of nation’s economy and at the same time citizens have fulfilled their dream to have a home for peaceful life in Indian context. The study aims to examine the reasons for sudden hike of properties we which has made it far from the common man. Sampling frame is pune construction company which are 20 in numbers. Both primary and secondary data has been used in the study. Introduction Talegaon Dabhade is a significant place in PUNE city; it’s a village with a municipal council, in Mawal- Taluka wherein team of BJP ward commissioners is in power. This place is even 25 km from Lonavla and 35 km from Pune. It is situated on highest altitude between the two metros (Mumbai and Pune) which are at 2200 feet above sea level. Talegaon is higher in altitude than the famous nearby hill stationlike Khandala and Lonavla (2047 ft.), thus it has pleasant weather throughout the year. Mainly, residentialproperty contains two type of structure: First, Structure for a single family and Second, Structure for a for multifamily structure. In Talegaon dabhade area mainly structure for multifamily structure are growing in full swing.Under these two types of structures, there are categories e.g. -

The Eruptive Tempo of Deccan Volcanism in Relation to The
RESEARCH 40 39 MASS EXTINCTION We measured 19 high-precision Ar/ Ar ages for the DT that complement the previous geo- chronology to create a higher-resolution tempo- ral framework for DT volcanism. We collected The eruptive tempo of Deccan samples for geochronological analysis from the Western Ghats region of the DT, the most rele- volcanism in relation to the vant region for understanding DT-induced climate change, as the record of the most voluminous Cretaceous-Paleogene boundary eruptive phase of the DT occurs here (Fig. 1). The total lava stratigraphy is called the Deccan Group, 1,2 1,3 4 5 which is divided into formations within the larger Courtney J. Sprain *, Paul R. Renne , Loÿc Vanderkluysen , Kanchan Pande , Stephen Self1, Tushar Mittal1 Kalsubai, Lonavala, and Wai subgroups (in ascend- ing order) (Fig. 1). These formational and subgroup boundaries arise from geochemical and volcano- Late Cretaceous records of environmental change suggest that Deccan Traps logical properties (23). Each formation comprises (DT) volcanism contributed to the Cretaceous-Paleogene boundary (KPB) multiple eruptive units. In total, we sampled each ecosystem crisis. However, testing this hypothesis requires identification of subgroup and all but two formations within these the KPB in the DT. We constrain the location of the KPB with high-precision subgroups, including the stratigraphically high- argon-40/argon-39 data to be coincident with changes in the magmatic plumbing est and lowest dated samples from the Western system. We also found that the DT did not erupt in three discrete large pulses and Ghats. Our samples came from multiple sections. -

Village Map Taluka: Khandala District: Satara
Village Map Taluka: Khandala District: Satara Bhor Purandhar Hartali Bhatghar Wadwadi µ Shindewadi Wing Bhatghar Reservoir 3 1.5 0 3 6 9 Rajewadi Shirwal km Pisalwadi Guthalwadi Moh tarf shirwal Mane Colony (N.V.) Tondal Location Index Palashi Sangvi Mirje Wathar Bk. Bholi Loni Baramati Dhangarwadi Vir Reservoir Kanhavadi Rui District Index Nandurbar Atit Shekhmirwadi Bhandara Vadgaon Naigaon Shedgewadi Dhule Amravati Nagpur Gondiya Bhade Pimpare Bk. Jalgaon Akola Wardha Buldana Bhadavade Andori Bavkalwadi Shivajinagar Nashik Washim Chandrapur Kesurdi Yavatmal Javale Aurangabad Padegaon Palghar Jalna Hingoli Gadchiroli Kavathe Bavda Karnavadi Thane Ahmednagar Parbhani Mariachiwadi Mumbai Suburban Nanded Morve Mumbai Bid Karadwadi Balu patlachiwadi Pune Pargaon Raigarh Bidar Waghoshi Latur Lohom Ghadgewadi Osmanabad Solapur Ajnuj KHANDALA Satara !( Mhavashi Ahire Zagalwadi Ratnagiri Asawali Sangli Kanheri Lonand (CT) Maharashtra State Limachiwadi Kolhapur Sindhudurg Ambarwadi Khandala Khed Bk. Dharwad Dhawadwadi Sukhed Wanyachiwadi Ghatdare Taluka Index Harali Khandala Bori Nimbodi Wai Phaltan Tambe Dharan Koregaon Yelewadi (N.V.) Mahabaleshwar Jaoli Man Padali Satara Phaltan Khatav Legend Koparde Patan !( Taluka Head Quarter Karad !( Wai Railway District: Satara Express Highway National Highway Koregaon Village maps from Land Record Department, GoM. Data Source: Waterbody/River from Satellite Imagery. District Boundary Taluka Boundary Generated By: Maharashtra Remote Sensing Applications Centre Village Boundary Autonomous Body of Planning Department, Waterbody/River Government of Maharashtra, VNIT Campus, Jaoli South Am bazari Road, Nagpur 440 010. -

Vaarivana Brochure Mumbai.Pdf
Vaarivana (Forest of Clouds) Welcome to Vaarivana- Pune’s largest first home villa community. It is time to move away from the life of concrete slabs, closed doors and safety frills. It’s time to embrace a new way of living. Vaarivana is designed to offer a lifestyle that’s unmatched by anything. A community created to let humans and nature coexist in harmony. And most importantly, a township that offers a safe, contemporary and healthy environment to let you enjoy the best that life has to offer. Artist’s impression Clear skies, open spaces, fresh air, stunning vistas and lots of beautiful memories. Artist’s impression Artist’s impression Vaarivana means a forest of clouds; which this really is. Nestled in the stunning landscape of Urse, Vaarivana is a distinctive low-rise development. It will feature only independent villas and twin villas, sitting cosily below the tree tops. All homes come with their private plots giving you the freedom to design the space you can create memories in. Image for representation only s 4 bedroom independent villas and 3 bedroom twin villas s Double height living room s Private terrace s Garden along the periphery s Optional private plunge pool s Around 70% of total space to be developed as open/green area s Aesthetically pleasing, environmentally sensitive and efficiently purposeful design s Every home with a unique view, with a hill on one side and a sloping landscape on the other Image for representation only Artist’s impression Artist’s impression 247 acres of pure bliss. Vaarivana is designed to help you lead a healthy, enriched and truly meaningful life. -

Description of the Region (Geographical Extent, Topography, Climate, and Vegetation)
Description of the Region (Geographical extent, topography, climate, and vegetation) The Maharashtra state is about 800 km east-west and 700 km north-south, an irregular dentate pentagon, lying between 22" r-16 " 4' north latitude and 72 " 6'-80 " 9' east longitude, covering an area of 3,07,690 sq km. It is limited to the west by the Arabian Sea, making a long coastline of 720 km. by Goa and Karnataka to the south, by Andhra Pradesh on the south-east, and Madhya Pradesh on the north, and Gujarat to its north-west (Map 1). Western Ghats or Sahyadri separate coastal strip of Konkan from rest of the plateau and thereby altitude ranges from mean sea level to about 1200 m on Western Ghats (with some highest peaks in the range like Kalsubai- 1654 m, Mahabaleshwar- 1382 m) and about 200-900 m over the rest. Average rainfall in the state varies from 250 cm in Konkan to 60-75 cm in Marathwada and again increasing to 150 cm towards eastern most part of Maharashtra that is Vidarbha. It forms a large part of Indian Peninsula. Similarly temperature varies between I5"C-47''C. Relative humidity fluctuate between 15% to 90%. Nearly 21% of the geographical area is under forest. Physiography Physiographically the state is divided into 5 divisions 1. Konkan, 2. Deccan or Desh, 3. Khandesh, 4. Marathwada and 5. Vidarbha (Map 2). Konkan, a narrow coastal strip of the west of Sahyadris, varies between 27-48 km in breadth and 800 km in length from Goa to Tapi Basin.