The Use of Environmental DNA for the Detection of Palaemonias Ganteri (Hay, 1901), a Federally Endangered Cave Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Eastslope Sculpin (Cottus Sp.) in Alberta
COSEWIC Assessment and Status Report on the "Eastslope" Sculpin Cottus sp. in Canada St. Mary and Milk River populations THREATENED 2005 COSEWIC COSEPAC COMMITTEE ON THE STATUS OF COMITÉ SUR LA SITUATION ENDANGERED WILDLIFE DES ESPÈCES EN PÉRIL IN CANADA AU CANADA COSEWIC status reports are working documents used in assigning the status of wildlife species suspected of being at risk. This report may be cited as follows: COSEWIC 2005. COSEWIC assessment and status report on the "eastslope" sculpin (St. Mary and Milk River population) Cottus sp. in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. vi + 30 pp. (www.sararegistry.gc.ca/status/status_e.cfm). Production note: This document is based on a report by Susan M. Pollard prepared for Alberta Sustainable Resource Development, Fish and Wildlife Division and the Alberta Conservation Association. The original report was published as Alberta Wildlife Status Report No. 51, February 2004, and is entitled Status of the St. Mary Shorthead Sculpin (provisionally Cottus bairdi punctulatus) in Alberta. Funding for the preparation of the original status report was provided by the Alberta Conservation Association and the Fish and Wildlife Division of Alberta Sustainable Resource Development. This document was overseen and edited by Bob Campbell, the COSEWIC Freshwater Fish Species Specialist Subcommittee Co- chair. For additional copies contact: COSEWIC Secretariat c/o Canadian Wildlife Service Environment Canada Ottawa, ON K1A 0H3 Tel.: (819) 997-4991 / (819) 953-3215 Fax: (819) 994-3684 E-mail: COSEWIC/[email protected] http://www.cosewic.gc.ca Ếgalement disponible en français sous le titre Ếvaluation et Rapport de situation du COSEPAC sur le chabot du versant est (populations des rivières St. -

Two Freshwater Shrimp Species of the Genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the Description of a New Species
A peer-reviewed open-access journal ZooKeys 923: 15–32 (2020) Caridina tetrazona 15 doi: 10.3897/zookeys.923.48593 RESEarcH articLE http://zookeys.pensoft.net Launched to accelerate biodiversity research Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species Qing-Hua Chen1, Wen-Jian Chen2, Xiao-Zhuang Zheng2, Zhao-Liang Guo2 1 South China Institute of Environmental Sciences, Ministry of Ecology and Environment, Guangzhou 510520, Guangdong Province, China 2 Department of Animal Science, School of Life Science and Enginee- ring, Foshan University, Foshan 528231, Guangdong Province, China Corresponding author: Zhao-Liang Guo ([email protected]) Academic editor: I.S. Wehrtmann | Received 19 November 2019 | Accepted 7 February 2020 | Published 1 April 2020 http://zoobank.org/138A88CC-DF41-437A-BA1A-CB93E3E36D62 Citation: Chen Q-H, Chen W-J, Zheng X-Z, Guo Z-L (2020) Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species. ZooKeys 923: 15–32. https://doi.org/10.3897/zookeys.923.48593 Abstract A faunistic and ecological survey was conducted to document the diversity of freshwater atyid shrimps of Dawanshan Island. Two species of Caridina that occur on this island were documented and discussed. One of these, Caridina tetrazona sp. nov. is described and illustrated as new to science. It can be easily distinguished from its congeners based on a combination of characters, which includes a short rostrum, the shape of the endopod of the male first pleopod, the segmental ratios of antennular peduncle and third maxilliped, the slender scaphocerite, and the absence of a median projection on the posterior margin. -

Fish Inventory at Stones River National Battlefield
Fish Inventory at Stones River National Battlefield Submitted to: Department of the Interior National Park Service Cumberland Piedmont Network By Dennis Mullen Professor of Biology Department of Biology Middle Tennessee State University Murfreesboro, TN 37132 September 2006 Striped Shiner (Luxilus chrysocephalus) – nuptial male From Lytle Creek at Fortress Rosecrans Photograph by D. Mullen Table of Contents List of Tables……………………………………………………………………….iii List of Figures………………………………………………………………………iv List of Appendices…………………………………………………………………..v Executive Summary…………………………………………………………………1 Introduction…………………………………………………………………...……..2 Methods……………………………………………………………………………...3 Results……………………………………………………………………………….7 Discussion………………………………………………………………………….10 Conclusions………………………………………………………………………...14 Literature Cited…………………………………………………………………….15 ii List of Tables Table1: Location and physical characteristics (during September 2006, and only for the riverine sites) of sample sites for the STRI fish inventory………………………………17 Table 2: Biotic Integrity classes used in assessing fish communities along with general descriptions of their attributes (Karr et al. 1986) ………………………………………18 Table 3: List of fishes potentially occurring in aquatic habitats in and around Stones River National Battlefield………………………………………………………………..19 Table 4: Fish species list (by site) of aquatic habitats at STRI (October 2004 – August 2006). MF = McFadden’s Ford, KP = King Pond, RB = Redoubt Brannan, UP = Unnamed Pond at Redoubt Brannan, LC = Lytle Creek at Fortress Rosecrans……...….22 Table 5: Fish Species Richness estimates for the 3 riverine reaches of STRI and a composite estimate for STRI as a whole…………………………………………………24 Table 6: Index of Biotic Integrity (IBI) scores for three stream reaches at Stones River National Battlefield during August 2005………………………………………………...25 Table 7: Temperature and water chemistry of four of the STRI sample sites for each sampling date…………………………………………………………………………….26 Table 8 : Total length estimates of specific habitat types at each riverine sample site. -
W7192e19.Pdf
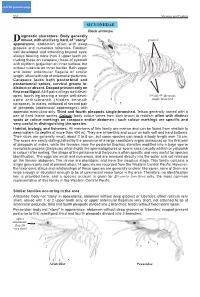
click for previous page 952 Shrimps and Prawns Sicyoniidae SICYONIIDAE Rock shrimps iagnostic characters: Body generally Drobust, with shell very hard, of “stony” grooves appearance; abdomen often with deep grooves and numerous tubercles. Rostrum well developed and extending beyond eyes, always bearing more than 3 upper teeth (in- cluding those on carapace); base of eyestalk with styliform projection on inner surface, but without tubercle on inner border. Both upper and lower antennular flagella of similar length, attached to tip of antennular peduncle. 1 Carapace lacks both postorbital and postantennal spines, cervical groove in- distinct or absent. Exopod present only on first maxilliped. All 5 pairs of legs well devel- 2 oped, fourth leg bearing a single well-devel- 3rd and 4th pleopods 4 single-branched oped arthrobranch (hidden beneath 3 carapace). In males, endopod of second pair 5 of pleopods (abdominal appendages) with appendix masculina only. Third and fourth pleopods single-branched. Telson generally armed with a pair of fixed lateral spines. Colour: body colour varies from dark brown to reddish; often with distinct spots or colour markings on carapace and/or abdomen - such colour markings are specific and very useful in distinguishing the species. Habitat, biology, and fisheries: All members of this family are marine and can be found from shallow to deep waters (to depths of more than 400 m). They are all benthic and occur on both soft and hard bottoms. Their sizes are generally small, about 2 to 8 cm, but some species can reach a body length over 15 cm. The sexes are easily distinguished by the presence of a large copulatory organ (petasma) on the first pair of pleopods of males, while the females have the posterior thoracic sternites modified into a large sperm receptacle process (thelycum) which holds the spermatophores or sperm sacs (usually whitish or yellowish in colour) after mating. -

Information on the NCWRC's Scientific Council of Fishes Rare
A Summary of the 2010 Reevaluation of Status Listings for Jeopardized Freshwater Fishes in North Carolina Submitted by Bryn H. Tracy North Carolina Division of Water Resources North Carolina Department of Environment and Natural Resources Raleigh, NC On behalf of the NCWRC’s Scientific Council of Fishes November 01, 2014 Bigeye Jumprock, Scartomyzon (Moxostoma) ariommum, State Threatened Photograph by Noel Burkhead and Robert Jenkins, courtesy of the Virginia Division of Game and Inland Fisheries and the Southeastern Fishes Council (http://www.sefishescouncil.org/). Table of Contents Page Introduction......................................................................................................................................... 3 2010 Reevaluation of Status Listings for Jeopardized Freshwater Fishes In North Carolina ........... 4 Summaries from the 2010 Reevaluation of Status Listings for Jeopardized Freshwater Fishes in North Carolina .......................................................................................................................... 12 Recent Activities of NCWRC’s Scientific Council of Fishes .................................................. 13 North Carolina’s Imperiled Fish Fauna, Part I, Ohio Lamprey .............................................. 14 North Carolina’s Imperiled Fish Fauna, Part II, “Atlantic” Highfin Carpsucker ...................... 17 North Carolina’s Imperiled Fish Fauna, Part III, Tennessee Darter ...................................... 20 North Carolina’s Imperiled Fish Fauna, Part -

Endangered Species
FEATURE: ENDANGERED SPECIES Conservation Status of Imperiled North American Freshwater and Diadromous Fishes ABSTRACT: This is the third compilation of imperiled (i.e., endangered, threatened, vulnerable) plus extinct freshwater and diadromous fishes of North America prepared by the American Fisheries Society’s Endangered Species Committee. Since the last revision in 1989, imperilment of inland fishes has increased substantially. This list includes 700 extant taxa representing 133 genera and 36 families, a 92% increase over the 364 listed in 1989. The increase reflects the addition of distinct populations, previously non-imperiled fishes, and recently described or discovered taxa. Approximately 39% of described fish species of the continent are imperiled. There are 230 vulnerable, 190 threatened, and 280 endangered extant taxa, and 61 taxa presumed extinct or extirpated from nature. Of those that were imperiled in 1989, most (89%) are the same or worse in conservation status; only 6% have improved in status, and 5% were delisted for various reasons. Habitat degradation and nonindigenous species are the main threats to at-risk fishes, many of which are restricted to small ranges. Documenting the diversity and status of rare fishes is a critical step in identifying and implementing appropriate actions necessary for their protection and management. Howard L. Jelks, Frank McCormick, Stephen J. Walsh, Joseph S. Nelson, Noel M. Burkhead, Steven P. Platania, Salvador Contreras-Balderas, Brady A. Porter, Edmundo Díaz-Pardo, Claude B. Renaud, Dean A. Hendrickson, Juan Jacobo Schmitter-Soto, John Lyons, Eric B. Taylor, and Nicholas E. Mandrak, Melvin L. Warren, Jr. Jelks, Walsh, and Burkhead are research McCormick is a biologist with the biologists with the U.S. -

ECOLOGY of NORTH AMERICAN FRESHWATER FISHES
ECOLOGY of NORTH AMERICAN FRESHWATER FISHES Tables STEPHEN T. ROSS University of California Press Berkeley Los Angeles London © 2013 by The Regents of the University of California ISBN 978-0-520-24945-5 uucp-ross-book-color.indbcp-ross-book-color.indb 1 44/5/13/5/13 88:34:34 AAMM uucp-ross-book-color.indbcp-ross-book-color.indb 2 44/5/13/5/13 88:34:34 AAMM TABLE 1.1 Families Composing 95% of North American Freshwater Fish Species Ranked by the Number of Native Species Number Cumulative Family of species percent Cyprinidae 297 28 Percidae 186 45 Catostomidae 71 51 Poeciliidae 69 58 Ictaluridae 46 62 Goodeidae 45 66 Atherinopsidae 39 70 Salmonidae 38 74 Cyprinodontidae 35 77 Fundulidae 34 80 Centrarchidae 31 83 Cottidae 30 86 Petromyzontidae 21 88 Cichlidae 16 89 Clupeidae 10 90 Eleotridae 10 91 Acipenseridae 8 92 Osmeridae 6 92 Elassomatidae 6 93 Gobiidae 6 93 Amblyopsidae 6 94 Pimelodidae 6 94 Gasterosteidae 5 95 source: Compiled primarily from Mayden (1992), Nelson et al. (2004), and Miller and Norris (2005). uucp-ross-book-color.indbcp-ross-book-color.indb 3 44/5/13/5/13 88:34:34 AAMM TABLE 3.1 Biogeographic Relationships of Species from a Sample of Fishes from the Ouachita River, Arkansas, at the Confl uence with the Little Missouri River (Ross, pers. observ.) Origin/ Pre- Pleistocene Taxa distribution Source Highland Stoneroller, Campostoma spadiceum 2 Mayden 1987a; Blum et al. 2008; Cashner et al. 2010 Blacktail Shiner, Cyprinella venusta 3 Mayden 1987a Steelcolor Shiner, Cyprinella whipplei 1 Mayden 1987a Redfi n Shiner, Lythrurus umbratilis 4 Mayden 1987a Bigeye Shiner, Notropis boops 1 Wiley and Mayden 1985; Mayden 1987a Bullhead Minnow, Pimephales vigilax 4 Mayden 1987a Mountain Madtom, Noturus eleutherus 2a Mayden 1985, 1987a Creole Darter, Etheostoma collettei 2a Mayden 1985 Orangebelly Darter, Etheostoma radiosum 2a Page 1983; Mayden 1985, 1987a Speckled Darter, Etheostoma stigmaeum 3 Page 1983; Simon 1997 Redspot Darter, Etheostoma artesiae 3 Mayden 1985; Piller et al. -

Chinese Mitten Crab (Eriocheir Sinensis) in San Francisco Bay
Distribution, Ecology and Potential Impacts of the Chinese Mitten Crab (Eriocheir sinensis) in San Francisco Bay Deborah A Rudnick Kathleen M. Halat Vincent H. Resh Department of Environmental Science, Policy and Management University of California, Berkeley TECHNICAL COMPLETION REPORT Project Number: UCAL-WRC-W-881 University of California Water Resources Center Contribution #206 ISBN 1-887192-12-3 June 2000 The University of California prohibits discrimination against or harassment of any person employed by or seeking employment with the University on the basis of race, color, national origin, religion, sex, physical or mental disability, medical condition (cancer- related), ancestry, marital status, age, sexual orientation, citizenship or status as a Vietnam-era veteran or special disabled veteran. The University of California is an affirmative action/equal opportunity employer. The University undertakes affirmative action to assure equal employment opportunity for underutilized minorities and women, for persons with disabilities, and for Vietnam-era veterans and special disabled veterans. University policy is intended to be consistent with the provisions of applicable State and Federal law. Inquiries regarding this policy may be addressed to the Affirmative Action Director, University of California, Agriculture and Natural Resources, 300 Lakeside Drive, 6th Floor, Oakland, CA 94612-3560, (510) 987-0097. This publication is a continuation in the Water Resources Center Contribution series. It is published and distributed by the UNIVERSITY -

LATE MIOCENE FISHES of the CACHE VALLEY MEMBER, SALT LAKE FORMATION, UTAH and IDAHO By
LATE MIOCENE FISHES OF THE CACHE VALLEY MEMBER, SALT LAKE FORMATION, UTAH AND IDAHO by PATRICK H. MCCLELLAN AND GERALD R. SMITH MISCELLANEOUS PUBLICATIONS MUSEUM OF ZOOLOGY, UNIVERSITY OF MICHIGAN, 208 Ann Arbor, December 17, 2020 ISSN 0076-8405 P U B L I C A T I O N S O F T H E MUSEUM OF ZOOLOGY, UNIVERSITY OF MICHIGAN NO. 208 GERALD SMITH, Editor The publications of the Museum of Zoology, The University of Michigan, consist primarily of two series—the Miscellaneous Publications and the Occasional Papers. Both series were founded by Dr. Bryant Walker, Mr. Bradshaw H. Swales, and Dr. W. W. Newcomb. Occasionally the Museum publishes contributions outside of these series. Beginning in 1990 these are titled Special Publications and Circulars and each is sequentially numbered. All submitted manuscripts to any of the Museum’s publications receive external peer review. The Occasional Papers, begun in 1913, serve as a medium for original studies based principally upon the collections in the Museum. They are issued separately. When a sufficient number of pages has been printed to make a volume, a title page, table of contents, and an index are supplied to libraries and individuals on the mailing list for the series. The Miscellaneous Publications, initiated in 1916, include monographic studies, papers on field and museum techniques, and other contributions not within the scope of the Occasional Papers, and are published separately. Each number has a title page and, when necessary, a table of contents. A complete list of publications on Mammals, Birds, Reptiles and Amphibians, Fishes, I nsects, Mollusks, and other topics is available. -

Cave Biodiversity of the Southern Cumberland Plateau Kirk S
b-3-guidebook_Guidebook3 6/18/2014 10:01 PM Page 159 Cave Biodiversity of the Southern Cumberland Plateau Kirk S. Zigler, NSS 62696; Matthew L. Niemiller, NSS 53235; and Danté B. Fenolio The South Cumberland Region of Tennessee, Alabama, and Georgia (Figure 1) is known for its tremendous diversity of caves, including huge pits, massive stream passages, and tight crawls. Less well known is that the region also supports tremendous cave biodiversity (Niemiller, Zigler, and Fenolio, 2013). Here we discuss many of the species that inhabit caves of the region, focusing on the southern Cumberland Plateau. Cave Biodiversity Four ecological classes of organisms can be found in caves: trogloxenes, subtroglophiles, eutroglophiles, and troglobionts (Culver and Pipan, 2009). Trogloxenes are not typically found in caves and cannot persist there for long periods of time. They must either find their way back to the surface or ultimately perish. Subtroglophiles are commonly found in caves but are associated with surface habitats for at least part of their life cycle. Some are seasonal inhabitants of caves and others move back and forth from cave to surface habitats for feeding, such as cave-roosting bats, cave crickets, and Allegheny Woodrats (Neotoma magister). Eutroglophiles are commonly found underground but can be found in surface habitats. Unlike trogloxenes and subtroglophiles, eutroglophiles can complete their entire life cycle Figure 1 - The South Cumberland Region at the junction of underground. Examples include the Cave Salamander Tennessee, Alabama, and Georgia. Figure courtesy of Nick Hollingshead. (Eurycea lucifuga) and the Cave Orbweaver (Meta ovalis). Troglobionts are obligate, permanent residents of subterranean habitats. -

California Freshwater Shrimparesmall.Females Areusually (Corpus)
Endangered SpecU.S. Environmental iesFacts Protection Agency California Freshwater Shrimp Syncaris pacifica Description and Ecology Status Endangered, listed October 31, 1988. of Marin, Sonoma and Napa counties. They are still found in all three counties but in much fewer numbers and fewer Critical Habitat Not designated. streams. The distribution of the shrimp has been separated Appearance The California freshwater shrimp is a 10- into four drainage units: 1) tributary streams of the lower legged crustacean belonging to the atyid family. Of the three Russian River drainage, that flow westward to the Pacific other atyid members in North America, two are also listed Ocean, 2) coastal streams flowing westward directly into as endangered (Kentucky cave shrimp (Palaemonias ganteri) the Pacific Ocean, 3) streams draining into Tomales Bay, Female California freshwater shrimp with egg s, courtes y of Larr y Serpa and Alabama cave shrimp (Palaemonias alabamae)) and one and 4) streams flowing southward into San Pablo Bay. While which once inhabited coastal streams in California is believed California freshwater shrimp in laboratories can tolerate The California freshwater shrimp to be extinct (Pasadena shrimp (Syncaris pasadenae)). brackish waters, they are unable to tolerate ocean salinities. is an endangered species. These species were believed to have been isolated from a It is thought that the isolated streams that harbor these Endangered species are marine environment during the Jurassic period. They are shrimp were once connected, but later separated by geologic plants and animals that are anatomically distinguished from other shrimp by the length of uplift and rising sea level. in immediate danger of their pincer-like claws (chelae) and the bristles (setae) at the Habitat Within the low elevation-low gradient streams tips of the first two chelae. -

How Might Sea Level Change Affect Arthropod Biodiversity in Anchialine Caves: a Comparison of Remipedia and Atyidae Taxa (Arthropoda: Altocrustacea)
Org Divers Evol (2014) 14:225–235 DOI 10.1007/s13127-014-0167-5 ORIGINAL ARTICLE How might sea level change affect arthropod biodiversity in anchialine caves: a comparison of Remipedia and Atyidae taxa (Arthropoda: Altocrustacea) Monica M. Moritsch & M. Joey Pakes & David R. Lindberg Received: 28 July 2013 /Accepted: 1 February 2014 /Published online: 28 February 2014 # Gesellschaft für Biologische Systematik 2014 Abstract Sea level change influences biodiversity of endem- shoreline, we evaluated how species richness and phylogenet- ic cave fauna to varying degrees. In anchialine systems, a ic diversity change with distance from the present and historic marine layer flows under less saline layers, each with differing ocean. Remipede species richness and phylogenetic diversity associated fauna. We assess the role of present and historic declined rapidly with distance from the ocean. Ninety-five (last glacial maximum – 18,000 years ago) distance from the percent of the remipedes surveyed were located within 7 km ocean in determining species richness and phylogenetic diver- of the present ocean and 18 km of the historic ocean. Atyid sity patterns for two groups of anchialine crustaceans: the species richness and phylogenetic diversity declined more marine-restricted Remipedia and a subset of groundwater- slowly with distance from the ocean than that of remipedes. inhabiting atyid shrimp with greater tolerance for salinity Atyid shrimp were also distributed over a broader range: 95 % variation. We calculated species richness and phylogenetic were located within 100 km of the present ocean and 240 km diversity per cave based on records of remipede and atyid of the historic ocean.