A Practical Workflow for Making Anatomical Atlases for Biological Research
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Autodesk Entertainment Creation Suite
Autodesk Entertainment Creation Suite Top Reasons to Buy and Upgrade Access the power of the industry’s top 3D modeling and animation technology in one unbeatable software suite. Autodesk® Entertainment Creation Suite Options: Autodesk® Maya® Autodesk® 3ds Max® Entertainment Creation Suite 2010 includes: Entertainment Creation Suite 2010 includes: • Autodesk® Maya® 2010 software • Autodesk® 3ds Max® 2010 • Autodesk® MotionBuilder® 2010 software • Autodesk® MotionBuilder® 2010 software • Autodesk® Mudbox™ 2010 software • Autodesk® Mudbox™ 2010 software Comprehensive Creative Toolsets The Autodesk Entertainment Creation Suite offers an expansive range of artist-driven tools designed to handle tough production challenges. With a choice of either Autodesk Maya 2010 software or Autodesk 3ds Max 2010 software, you have access to award-winning, 3D software for modeling, animation, rendering, and effects. The Suite also includes Autodesk Mudbox 2010 software, allowing you to quickly and intuitively sculpt highly detailed models; and Autodesk MotionBuilder 2010 software, to quickly and efficiently create, manipulate and process massive amounts of animation data. The complementary toolsets of the Suite help you to achieve higher quality results more efficiently and more cost-effectively. Real-Time Performance with MotionBuilder The addition of MotionBuilder to a Maya or 3ds Max pipeline helps increase production efficiency, and produce higher quality results when developing projects requiring high-volume character animation. With its real-time 3D engine and dedicated toolsets for character rigging, nonlinear animation editing, motion-capture data manipulation, and interactive dynamics, MotionBuilder is an ideal, complementary toolset to Maya or 3ds Max, forming a unified Image courtesy of Wang Xiaoyu. end-to-end animation solution. Digital Sculpting and Texture Painting with Mudbox Designed by professional artists in the film, games and design industries, Mudbox software gives 3D modelers and texture artists the freedom to create without worrying about technical details. -

Multimedia Systems DCAP303
Multimedia Systems DCAP303 MULTIMEDIA SYSTEMS Copyright © 2013 Rajneesh Agrawal All rights reserved Produced & Printed by EXCEL BOOKS PRIVATE LIMITED A-45, Naraina, Phase-I, New Delhi-110028 for Lovely Professional University Phagwara CONTENTS Unit 1: Multimedia 1 Unit 2: Text 15 Unit 3: Sound 38 Unit 4: Image 60 Unit 5: Video 102 Unit 6: Hardware 130 Unit 7: Multimedia Software Tools 165 Unit 8: Fundamental of Animations 178 Unit 9: Working with Animation 197 Unit 10: 3D Modelling and Animation Tools 213 Unit 11: Compression 233 Unit 12: Image Format 247 Unit 13: Multimedia Tools for WWW 266 Unit 14: Designing for World Wide Web 279 SYLLABUS Multimedia Systems Objectives: To impart the skills needed to develop multimedia applications. Students will learn: z how to combine different media on a web application, z various audio and video formats, z multimedia software tools that helps in developing multimedia application. Sr. No. Topics 1. Multimedia: Meaning and its usage, Stages of a Multimedia Project & Multimedia Skills required in a team 2. Text: Fonts & Faces, Using Text in Multimedia, Font Editing & Design Tools, Hypermedia & Hypertext. 3. Sound: Multimedia System Sounds, Digital Audio, MIDI Audio, Audio File Formats, MIDI vs Digital Audio, Audio CD Playback. Audio Recording. Voice Recognition & Response. 4. Images: Still Images – Bitmaps, Vector Drawing, 3D Drawing & rendering, Natural Light & Colors, Computerized Colors, Color Palletes, Image File Formats, Macintosh & Windows Formats, Cross – Platform format. 5. Animation: Principle of Animations. Animation Techniques, Animation File Formats. 6. Video: How Video Works, Broadcast Video Standards: NTSC, PAL, SECAM, ATSC DTV, Analog Video, Digital Video, Digital Video Standards – ATSC, DVB, ISDB, Video recording & Shooting Videos, Video Editing, Optimizing Video files for CD-ROM, Digital display standards. -

Texture Mapping with Mudbox and 3Ds Max
Texture Mapping with Mudbox and 3ds Max by Joshua Holland Graphic Communication Department College of Liberal Arts California Polytechnic State University 2011 ABSTRACT Texture Mapping with Mudbox and 3ds Max Joshua Holland Graphic Communication Department, December 2011 Advisor: Kevin Cooper The purpose of this study was to determine the intuitiveness of texture mapping and compressibility of files generated using Autodesk 3ds Max 2012 versus Autodesk Mudbox 2012. This will be used by anyone starting to learn how to texture map and who is comparing programs that have the capability to do so. This will save users time in researching which program is better suited for their needs. This study investigated how Autodesk 3ds Max 2012 and Autodesk Mudbox 2012 compared in mapping textures to 3D models. A basic computer skills assessment test and an intuitive test was administered to twelve participants. They were ranked based on their computer skills assessment scores, texture map completion time with 3ds Max, and texture map completion time with Mudbox. Exported files sizes for 3D models using 3ds Max and Mudbox were also compared to determine which was smaller. Participants were not included in the file size portion of the study. Results from twelve participants in the intuitive test showed that Mudbox proved to be more intuitive for first time users attempting to apply a texture to a 3D object. However, 3ds Max offers more control and precision in performing these tasks at the cost of a large learning curve and a less of a fluid interface. The compressiblity test showed that 3ds Max produced a smaller file size. -
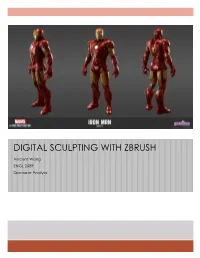
Digital Sculpting with Zbrush
DIGITAL SCULPTING WITH ZBRUSH Vincent Wang ENGL 2089 Discourse Analysis 2 ZBrush Analysis Table of Contents Context ........................................................................................................................... 3 Process ........................................................................................................................... 5 Analysis ........................................................................................................................ 13 Application .................................................................................................................. 27 Activity .......................................................................................................................... 32 Works Cited .................................................................................................................. 35 3 Context ZBrush was created by the Pixologic Inc., which was founded by Ofer Alon and Jack Rimokh (Graphics). It was first presented in 1999 at SIGGRAPH (Graphics). Version 1.5 was unveiled at the MacWorld Expo 2002 in New York and SIGGRAPH 2002 in San Antonio (Graphics). Pixologic, the company describes the 3D modeling software as a “digital sculpting and painting program that has revolutionized the 3D industry…” (Pixologic). It utilizes familiar real-world tools in a digital environment, getting rid of steep learning curves and allowing the user to be freely creative instead of figuring out all the technical details. 3D models that are created in -

HP and Autodesk Create Stunning Digital Media and Entertainment with HP Workstations
HP and Autodesk Create stunning digital media and entertainment with HP Workstations. Does your workstation meet your digital Performance: Advanced compute and visualization power help speed your work, beat deadlines, and meet expectations. At the heart of media challenges? HP Z Workstations are the new Intel® processors with advanced processor performance technologies and NVIDIA Quadro professional It’s no secret that the media and entertainment industry is constantly graphics cards with the NVIDIA CUDA parallel processing architecture; evolving, and the push to deliver better content faster is an everyday delivering real-time previewing and editing of native, high-resolution challenge. To meet those demands, technology matters—a lot. You footage, including multiple layers of 4K video. Intel® Turbo Boost1 need innovative, high-performing, reliable hardware and software tools is designed to enhance the base operating frequency of processor tuned to your applications so your team can create captivating content, cores, providing more processing speed for single and multi-threaded meet tight production schedules, and stay on budget. HP offers an applications. The HP Z Workstation cooling design enhances this expansive portfolio of integrated workstation hardware and software performance. solutions designed to maximize the creative capabilities of Autodesk® software. Together, HP and Autodesk help you create stunning digital Reliability: HP product testing includes application performance, media. graphics and comprehensive ISV certification for maximum productivity. All HP Workstations come with a limited 3-year parts, 3-year labor and The HP Difference 3-year onsite service (3/3/3) standard warranty that is extendable up to 5 years.2 You can be confident in your HP and Autodesk solution. -

PELC253 Digital Sculpting with Zbrush 2020-21.Docx
Glasgow School of Art Course Specification Course Title: Digital Sculpting with ZBrush Course Specifications for 2020/21 have not been altered in response to the COVID-19 pandemic. Please refer to the 2020/21 Programme Specification, the relevant Canvas pages and handbook for the most up-to-date information regarding any changes to a course. Course Code: HECOS Code: Academic Session: PELC253 2020-21 1. Course Title: Digital Sculpting with ZBrush 2. Date of Approval: 3. Lead School: 4. Other Schools: PACAAG April 2020 School of Simulation and This course is available to Visualisation students on PGT programmes which include a Stage 2 elective. 5. Credits: 6. SCQF Level: 7. Course Leader: 20 11 Dr. Sandy Louchart 8. Associated Programmes: This course is available to students on PGT programmes which include a Stage 2 elective. 9. When Taught: Semester 2 10. Course Aims: The overarching aims of the cross-school electives are to: • Encourage interdisciplinary, critical reflexivity from within an open set of choices; • Foster deep investigative approaches to new or unfamiliar areas of practice and theory; • Cultivate self-directed leadership and initiative-taking in both applied and abstract modes of • practice/ study not necessarily associated with a student’s particular creative specialism; • Enable flexible, ethical exploration and connection of diverse knowledge and understanding • within a specialist programme of study. The practice-based and skill focussed course provides a thorough and intensive introduction to digital 3D sculpting, allowing students to obtain a high-level of proficiency in this technically challenge discipline. Students will work with a range of techniques and practices through which a digital painting can be produced and distributed. -

3D Modeling and the Role of 3D Modeling in Our Life
ISSN 2413-1032 COMPUTER SCIENCE 3D MODELING AND THE ROLE OF 3D MODELING IN OUR LIFE 1Beknazarova Saida Safibullaevna 2Maxammadjonov Maxammadjon Alisher o’g’li 2Ibodullayev Sardor Nasriddin o’g’li 1Uzbekistan, Tashkent, Tashkent University of Informational Technologies, Senior Teacher 2Uzbekistan, Tashkent, Tashkent University of Informational Technologies, student Abstract. In 3D computer graphics, 3D modeling is the process of developing a mathematical representation of any three-dimensional surface of an object (either inanimate or living) via specialized software. The product is called a 3D model. It can be displayed as a two-dimensional image through a process called 3D rendering or used in a computer simulation of physical phenomena. The model can also be physically created using 3D printing devices. Models may be created automatically or manually. The manual modeling process of preparing geometric data for 3D computer graphics is similar to plastic arts such as sculpting. 3D modeling software is a class of 3D computer graphics software used to produce 3D models. Individual programs of this class are called modeling applications or modelers. Key words: 3D, modeling, programming, unity, 3D programs. Nowadays 3D modeling impacts in every sphere of: computer programming, architecture and so on. Firstly, we will present basic information about 3D modeling. 3D models represent a physical body using a collection of points in 3D space, connected by various geometric entities such as triangles, lines, curved surfaces, etc. Being a collection of data (points and other information), 3D models can be created by hand, algorithmically (procedural modeling), or scanned. 3D models are widely used anywhere in 3D graphics. -

A Practical Workflow for Making Anatomical Atlases for Biological Research
A Practical Workflow for Making Anatomical Atlases for Biological Research Yong Wan, A. Kelsey Lewis, Mary Colasanto, Mark van Langeveld, Gabrielle Kardon, and Charles Hansen ■ University of Utah An anatomical atlas provides a detailed map for medical and biological studies of anatomy. These atlases are important for understanding normal anatomy and the development and function of structures, and for determining the etiology of congenital abnormalities. Unfortunately, for biologists, generating such atlases is difficult, especially ones with the informative content and aesthetic quality that characterize human anatomy atlases. Building such atlases requires knowledge of the species being studied and experience with an art form that can faithfully record and present this knowledge, both of which require extensive training in considerably different fields. (For some background on anatomical atlases, see the related sidebar.) With the latest innovations in data acquisition and computing techniques, atlas building has changed dramatically. We can now create atlases from 3D images of biological specimens, allowing for high-quality, faithful representations. Labeling of structures using fluorescently tagged antibodies, confocal 3D scanning of these labeled structures, volume rendering, segmentation, and surface reconstruction techniques all promise solutions to the problem of building atlases. However, biology researchers still ask, “Is there a set of tools we can use or a practical workflow we can follow so that we can easily build models from our biological data?” To help answer this question, computer scientists have developed many algorithms, tools, and program codes. Unfortunately, most of these researchers have tackled only one aspect of the problem or provided solutions to special cases. So, the general question of how to build anatomical atlases remains unanswered. -

Digital Sculpting with Mudbox This Page Intentionally Left Blank Digital Sculpting with Mudbox
Digital Sculpting with Mudbox This page intentionally left blank Digital Sculpting with Mudbox Essential Tools and Techniques for Artists Mike de la Flor Bridgette Mongeon AMSTERDAM • BOSTON • HEIDELBERG • LONDON NEW YORK • OXFORD • PARIS • SAN DIEGO SAN FRANCISCO • SINGAPORE • SYDNEY • TOKYO Focal Press is an Imprint of Elsevier This book is dedicated to the community of artists and researchers whose creativity, hard work, and zeal for innovation push the technological envelopes to greater heights. Focal Press is an imprint of Elsevier 30 Corporate Drive, Suite 400, Burlington, MA 01803, USA The Boulevard, Langford Lane, Kidlington, Oxford, OX5 1GB, UK © 2010 Elsevier Inc. All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or any information storage and retrieval system, without permission in writing from the publisher. Details on how to seek permission, further information about the Publisher’s permissions policies and our arrangements with organizations such as the Copyright Clearance Center and the Copyright Licensing Agency, can be found at our website: www.elsevier.com/permissions. This book and the individual contributions contained in it are protected under copyright by the Publisher (other than as may be noted herein). Notices Knowledge and best practice in this field are constantly changing. As new research andexperience broaden our understanding, changes in research methods, professional practices, or medical treatment may become necessary. Practitioners and researchers must always rely on their own experience and knowledge in evaluating and using any information, methods, compounds, or experiments described herein. In using such information or methods they should be mindful of their own safety and the safety of others, including parties for whom they have a professional responsibility. -

Interactive Graphics
Interactive Graphics Prof. Marco Schaerf Dept. of Computer, Systems and Management Science (DIAG) Sapienza University of Rome [email protected] Interactive Graphics: Introduction 10/2/2014 Pagina 1 Plan for today • Syllabus • Logistics • Computer graphics Interactive Graphics: Introduction 10/2/2014 Pagina 2 Syllabus (Core) • Introduction, Color, Graphics pipeline • WebGL, 3D modeling, Transformation • Rasterization, Clipping • Lighting and shading • Texture mapping • Advanced Techniques, Global Illumination Interactive Graphics: Introduction 10/2/2014 Pagina 3 Syllabus (Optional) • Graphics hardware, intro to GPGPU • Animation introduction, particle systems, rigid bodies simulation • Topics in animation and modeling • Research topics Interactive Graphics: Introduction 10/2/2014 Pagina 4 Contents • you will • understand image synthesis principles • learn math to make images • implement key algorithms • write cool apps • learn graphics Javascript APIs (WebGL) • you will not • implement large systems Interactive Graphics: Introduction 10/2/2014 Pagina 5 Website, email, office hours • Website: https://piazza.com/uniroma1.it/fall2014/1044398/home • Email: [email protected] • Office hours: after each lesson or by appointment in room B220 Interactive Graphics: Introduction 10/2/2014 Pagina 6 Recommended Books • Fabio Ganovelli, Massimiliano Corsini, Sumanta Pattanaik, Marco Di Benedetto, Introduction to Computer Graphics: A Practical Learning Approach CRC Press (also available in e-book format) • Angel and Shreiner, Interactive -

Introducing Zbrush® ERIC KELLER
62795ffirs.qxd 3/25/08 7:18 PM Page iii Introducing ZBrush® ERIC KELLER WILEY PUBLISHING, INC. 62795ffirs.qxd 3/23/08 8:30 AM Page ii 62795ffirs.qxd 3/23/08 8:30 AM Page i Introducing ZBrush® 62795ffirs.qxd 3/23/08 8:30 AM Page ii 62795ffirs.qxd 3/25/08 7:18 PM Page iii Introducing ZBrush® ERIC KELLER WILEY PUBLISHING, INC. 62795ffirs.qxd 3/23/08 8:30 AM Page iv Acquisitions Editor: Mariann Barsolo Development Editor: Stephanie Barton Technical Editor: Gael McGill, PhD Production Editor: Rachel Gunn Copy Editor: Judy Flynn Production Manager: Tim Tate Vice President and Executive Group Publisher: Richard Swadley Vice President and Executive Publisher: Joseph B. Wikert Vice President and Publisher: Neil Edde Media Associate Project Manager: Laura Atkinson Media Assistant Producer: Josh Frank Media Quality Assurance: Kit Malone Book Designer: Caryl Gorska Compositors: Chris Gillespie, Kate Kaminski, Happenstance Type-O-Rama Proofreader: Ian Golder Indexer: Ted Laux Cover Designer: Ryan Sneed Cover Images: Eric Keller Copyright © 2008 by Wiley Publishing, Inc., Indianapolis, Indiana Published simultaneously in Canada ISBN: 978-0-470-26279-5 No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying, recording, scanning or otherwise, except as permitted under Sections 107 or 108 of the 1976 United States Copyright Act, without either the prior written permission of the Publisher, or authorization through payment of the appropriate per-copy fee to the Copyright Clearance Center, 222 Rosewood Drive, Danvers, MA 01923, (978) 750-8400, fax (978) 646-8600. -

Workflows for Creating 3D Game Characters
Tapio Terävä Workflows for Creating 3D Game Characters Bachelor of Business Administration Business Information Technology Spring 2017 TIIVISTELMÄ Tekijä: Tapio Terävä Työn nimi: Työtapoja 3D pelihahmojen luomiseen Tutkintonimike: Tradenomi (AMK), Tietojenkäsittely Asiasanat: peli, hahmo, pelihahmo, 3D, työtapa Opinnäytetyö käsittelee 3D pelihahmojen luomiseen käytettäviä erilaisia työtapoja, sekä niissä käy- tettäviä eri työkaluja. Lisäksi opinnäytetyö pyrkii luomaan kokonaisvaltaisen kuvan 3D pelihahmo- jen luomisprosessista, siihen liittyvistä keskeisistä teknologioista, sekä rajoituksista joita erilaiset pelialustat asettavat hahmojen luomiselle. Opinnäytetyö käsittelee alkuun pelihahmojen luomisen eri lähtökohtia, sekä hahmosuunnittelun eri käytäntöjä ja työtapoja. Seuraavaksi käydään läpi yleistetty esimerkki 3D pelihahmon luomispro- sessista, johon sisältyy useita eri työvaiheita. Yleistetyn esimerkin jälkeen esitellään tästä poik- keavia työtapoja, joiden eroavaisuudet johtuvat eri pelialustojen asettamista rajoitteista tai niiden tarjoamista mahdollisuuksista. Lopuksi vertaillaan miten perinteiset työtavat eroavat nykyaikaisista työtavoista, ja esitellään uusien työtapojen etuja. ABSTRACT Author: Tapio Terävä Title of the Publication: Workflows for Creating 3D Game Characters Degree Title: Bachelor of Business Administration (UAS), Business Information Technology Keywords: game, character, game character, 3D, workflow This thesis deals with the various workflows, methods, and tools used for creating 3D game char- acters. The objective of the thesis was to construct a complete picture of the creation process of 3D game characters, and the related technologies essential to the process, as well as the re- strictions set by different game platforms. First, the different motives for creating game characters are introduced, along with the different practices and methods used for designing characters. This is followed by a generalized example of the process of creating a 3D game character, which consists of several different stages.