Assignment 1 Part I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of NF-Κb and C/EBP Factors During Pathogen-Mediated
The role of NF-B and C/EBP factors during pathogen-mediated activation of bovine interleukin 8 and beta-defensin in mammary epithelial cells Inaugural dissertation for the academic degree Doctor rerum naturalium of Mathematisch-Naturwissenschaftlichen Fakultät Universität Rostock By Shuzhen Liu (M. Sc.), born on March-02-1972, in Shanxi Province, China From the Forschungsinstitut für die Biologie landwirtschaftlicher Nutztiere in Dummerstorf Rostock 2009 URN: urn:nbn:de:gbv:28-diss2009-0185-5 Dean: Prof. Dr. Hendrik Schubert Reviewers: 1. Prof. Dr. Hans-Martin Seyfert Research Unit molecular biology, Research Institute for the Biology of Farm Animals, Wilhelm-Stahl-Allee 2, D-18196 Dummerstorf, Germany 2. Prof. Dr. Dieter G. Weiss Division of animal Physiology, Institute of Cell Biology and Biosystems Technology, University of Rostock, Albert-Einstein-Strasse 3, 18059 Rostock, Germany 3. PD Dr. Ulrike Gimsa Research Unit Behavioural Physiology, Research Institute for the Biology of Farm Animals, Wilhelm-Stahl-Allee 2, D-18196 Dummerstorf, Germany Date of defense: October 19th, 2009 Table of Contents TABLE OF CONTENTS 1. INTRODUCTION..................................................................................................................1 1.1 Mastitis as a challenge in general immunology..................................................................1 1.2 Innate immunity of the bovine mammary gland ................................................................2 1.3 Toll-like receptors (TLRs): main receptors perceiving the pathogen -

Restriction Endonucleases
Molecular Biology Problem Solver: A Laboratory Guide. Edited by Alan S. Gerstein Copyright © 2001 by Wiley-Liss, Inc. ISBNs: 0-471-37972-7 (Paper); 0-471-22390-5 (Electronic) 9 Restriction Endonucleases Derek Robinson, Paul R. Walsh, and Joseph A. Bonventre Background Information . 226 Which Restriction Enzymes Are Commercially Available? . 226 Why Are Some Enzymes More Expensive Than Others? . 227 What Can You Do to Reduce the Cost of Working with Restriction Enzymes? . 228 If You Could Select among Several Restriction Enzymes for Your Application, What Criteria Should You Consider to Make the Most Appropriate Choice? . 229 What Are the General Properties of Restriction Endonucleases? . 232 What Insight Is Provided by a Restriction Enzyme’s Quality Control Data? . 233 How Stable Are Restriction Enzymes? . 236 How Stable Are Diluted Restriction Enzymes? . 236 Simple Digests . 236 How Should You Set up a Simple Restriction Digest? . 236 Is It Wise to Modify the Suggested Reaction Conditions? . 237 Complex Restriction Digestions . 239 How Can a Substrate Affect the Restriction Digest? . 239 Should You Alter the Reaction Volume and DNA Concentration? . 241 Double Digests: Simultaneous or Sequential? . 242 225 Genomic Digests . 244 When Preparing Genomic DNA for Southern Blotting, How Can You Determine If Complete Digestion Has Been Obtained? . 244 What Are Your Options If You Must Create Additional Rare or Unique Restriction Sites? . 247 Troubleshooting . 255 What Can Cause a Simple Restriction Digest to Fail? . 255 The Volume of Enzyme in the Vial Appears Very Low. Did Leakage Occur during Shipment? . 259 The Enzyme Shipment Sat on the Shipping Dock for Two Days. -

CLONING, EXPRESSION ANALYSIS, and TRANSFORMATION VECTOR CONSTRUCTION of DAM HOMOLOGS in PEACH and POPLAR Yuhui Xie Clemson University, [email protected]
Clemson University TigerPrints All Theses Theses 1-2011 CLONING, EXPRESSION ANALYSIS, AND TRANSFORMATION VECTOR CONSTRUCTION OF DAM HOMOLOGS IN PEACH AND POPLAR Yuhui Xie Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_theses Part of the Plant Biology Commons Recommended Citation Xie, Yuhui, "CLONING, EXPRESSION ANALYSIS, AND TRANSFORMATION VECTOR CONSTRUCTION OF DAM HOMOLOGS IN PEACH AND POPLAR" (2011). All Theses. 1146. https://tigerprints.clemson.edu/all_theses/1146 This Thesis is brought to you for free and open access by the Theses at TigerPrints. It has been accepted for inclusion in All Theses by an authorized administrator of TigerPrints. For more information, please contact [email protected]. CLONING, EXPRESSION ANALYSIS, AND TRANSFORMATION VECTOR CONSTRUCTION OF DAM HOMOLOGS IN PEACH AND POPLAR _______________________________________________ A Thesis Presented to the Graduate School of Clemson University ________________________________________________ In Partial Fulfillment of the Requirement for the Degree Master of Science Plant and Environmental Sciences _________________________________________________ by Yuhui Xie August 2011 ________________________________________________ Accepted by: Dr. Douglas G. Bielenberg, Committee Chair Dr. Haiying Liang Dr. Hong Luo ABSTRACT Genetic fine mapping and sequencing of the EVG locus in peach [Prunus persica (L.) Batsch] identified six tandem arrayed Dormancy-Associated MADS-box (DAM) genes as candidates for regulating growth cessation and terminal bud formation in the non-dormant evergrowing (evg) mutant. Since the mutant is lacking expression of six genes in the mapped locus, further functional analysis is needed to narrow the list of gene candidates for the non-dormant evg phenotype. Here I report three sets of experiments designed to functionally test DAM genes in peach and their homologs in a model tree, hybrid poplar. -

Enzymes in Cloning Part I
˹̀/˺̀/˺̊˼̊ ENZYMES IN CLONING PART I Dr.Sarookhani ˺ ˹̀/˺̀/˺̊˼̊ Cloning --aa definition •• From the Greek --klon,klon, a twig •• An aggregate of the asexually produced progeny of an individual;a group of replicas of all or part of a macromolecule (such as DNA or an antibody) •• An individual grown from a single somatic cell of its parent & genetically identical to it •• Clone: a collection of molecules or cells, all identical to an original molecule or cell Dr.Sarookhani ˻ ˹̀/˺̀/˺̊˼̊ Different types of Cloning 1. Reproductive Cloning 2. Therapeutic Cloning 3. Recombinant DNA Technology or DNA Cloning Dr.Sarookhani ˼ ˹̀/˺̀/˺̊˼̊ DNA CLONING A method for identifying and purifying a particular DNA fragment (clone) of interest from a complex mixture of DNA fragments, and then producing large numbers of the fragment (clone) of interest. Dr.Sarookhani ̊ ˹̀/˺̀/˺̊˼̊ What is genetic engineering • Genetic engineering, also known as recombinant DNA technology, means altering the genes in a living organism to produce a Genetically Modified Organism (GMO) with a new genotype. • Various kinds of genetic modification are possible: inserting a foreign gene from one species into another, forming a transgenic organism; altering an existing gene so that its product is changed; or changing gene expression so that it is translated more often or not at all. Dr.Sarookhani ̋ ˹̀/˺̀/˺̊˼̊ Dr.Sarookhani ̌ ˹̀/˺̀/˺̊˼̊ Dr.Sarookhani ̀ ˹̀/˺̀/˺̊˼̊ Genomic Library Dr.Sarookhani ́ ˹̀/˺̀/˺̊˼̊ Dr.Sarookhani ̂ ˹̀/˺̀/˺̊˼̊ Basic steps in genetic engineering 1. -

Cyclic Digestion and Ligation-Mediated PCR Used For
www.nature.com/scientificreports OPEN Cyclic Digestion and Ligation- Mediated PCR Used for Flanking Sequence Walking Dong Yu1,2,5, Tianshun Zhou2,4,5, Xuewu Sun2,3, Zhizhong Sun2, Xiabing Sheng1,2, Yanning Tan2, Ling Liu2,4, Ning Ouyang2,4, Ke Xu2, Kaibing Shi2, Guilong Yuan2, Jia Ding2, Meijuan Duan3* & Dingyang Yuan1,2,3,4* Ligation-mediated PCR (LM-PCR) is a classical method for isolating fanking sequences; however, it has a common limitation of reduced success rate owing to the circularization or multimerization of target restriction fragments including the known sequence. To address this limitation, we developed a novel LM-PCR method, termed Cyclic Digestion and Ligation-Mediated PCR (CDL-PCR). The novelty of this approach involves the design of new adapters that cannot be digested after being ligated with the restriction fragment, and cyclic digestion and ligation may be manipulated to block the circularization or multimerization of the target restriction fragments. Moreover, to improve the generality and fexibility of CDL-PCR, an adapter precursor sequence was designed, which could be digested to prepare 12 diferent adapters at low cost. Using this method, the fanking sequences of T-DNA insertions were obtained from transgenic rice and Arabidopsis thaliana. The experimental results demonstrated that CDL-PCR is an efcient and fexible method for identifying the fanking sequences in transgenic rice and Arabidopsis thaliana. Identifcation of fanking sequences has ofen been employed to determine the location of T-DNA insertion in genomic DNA. Methods to obtain fanking sequencea include inverse PCR1, randomly primed PCR2–5, and ligation-mediated PCR6–8. Inverse PCR, the earliest fanking cloning technique, has a low efciency and is limited by the rate of self-ligation and amplifcation range of DNA polymerases9. -

Bpuami: a Novel Saci Neoschizomer from Bacillus Pumilus Discovered in an Isolate from Amazon Basin, Recognizing 5'-Gag↓Ctc-3'
Brazilian Journal of Microbiology (2006) 37:96-100 ISSN 1517-8382 BPUAMI: A NOVEL SACI NEOSCHIZOMER FROM BACILLUS PUMILUS DISCOVERED IN AN ISOLATE FROM AMAZON BASIN, RECOGNIZING 5'-GAG↓CTC-3' Jocelei M. Chies1,2,*; Ana C. de O. Dias1; Hélio M. M. Maia3; Spartaco Astolfi-Filho2,3 1Centro de Biotecnologia, Universidade Federal do Rio Grande do Sul, RS, Brasil; 2Curso de Pós-Graduação em Biologia Molecular, Universidade de Brasília, Brasília, DF, Brasil; 3Centro de Apoio Multidisciplinar, Universidade Federal do Amazonas, Manaus, AM, Brasil Submitted: June 27, 2005; Returned to authors for corrections: November 16, 2005; Approved: January 12, 2006 ABSTRACT A strain of Bacillus pumilus was isolated and identified from water samples collected from a small affluent of the Amazon River. Type II restriction endonuclease activity was detected in these bacteria. The enzyme was purified and the molecular weight of the native protein estimated by gel filtration and SDS-PAGE. The optimum pH, temperature and salt requirements were determined. Quality control assays showed the complete absence of “nonspecific nucleases.” Restriction cleavage analysis and DNA sequencing of restriction fragments allowed the unequivocal demonstration of 5´GAG↓CTC3´ as the recognition sequence. This enzyme was named BpuAmI and is apparently a neoschizomer of the prototype restriction endonuclease SacI. This is the first report of an isoschizomer and/or neoschizomer of the prototype SacI identified in the genus Bacillus. Key words: type II restriction endonuclease, BpuAmI, SacI, neoschizomers, Bacillus pumilus INTRODUCTION thousands of taxonomically diverse bacteria for enzymes with new characteristics. However, to our knowledge, there has Restriction endonucleases are enzymes which recognize not been any report to date of an isoschizomer and/or short DNA sequences and cleave DNA in both strands. -

1. Restriction Endonucleases (® Fermentas 2006)
We guarantee that these products are Introduction ..........................................................................................................2 free of contaminating activities. Restriction Endonucleases Guide Our stringent quality control with the most FastDigest™ Restriction Endonucleases .........................................................2 Fermentas Restriction Endonucleases............................................................3 advanced tests guarantees you pure 1 Alphabetic List of Commercially Available Restriction Endonucleases.........6 products for your experiments. ISO9001 Recognition Specifi cities...............................................................................16 and ISO14001 is your assurance of con- Commercial Restriction Enzymes Sorted by the Type of DNA Ends Generated sistency and lot-to-lot reproducibility. Enzymes Generating 5’-protruding Ends.......................................................18 Enzymes Generating 3’-protruding Ends.......................................................18 ™ PureExtreme Quality will provide the Enzymes Generating Blunt Ends...................................................................19 performance you need for your most Enzymes Cleaving DNA on the Both Sides of Their Recognition Sequences.....19 demanding experiments. PureExtreme™ Quality PureExtreme™ Quality Guarantee..................................................................20 Activity Assay.................................................................................................20 -
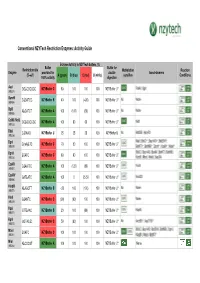
Conventional Nzytech Restriction Enzymes: Activity Guide
Conventional NZYTech Restriction Enzymes: Activity Guide Enzyme Activity in NZYTech buffers (%) Buffer Buffer for Restriction site Methylation Reaction Enzyme provided for double Isoschizomers (5’ →→→3’) A (green) B (blue) C (red) U (white) sensitive Conditions 100% activity digestion AscI GG ↓↓↓CGCGCC NZYBuffer C 80 100 100 100 NZYBuffer U * PalAI, SgsI (MB231) BamHI G↓↓↓GATCC NZYBuffer B 40 100 (<20) 100 NZYBuffer U * No None (MB064) BglII A↓↓↓GATCT NZYBuffer A 100 (100) (60) 100 NZYBuffer U * No None (MB065) CciNI (NotI) GC ↓↓↓GGCCGC NZYBuffer A 100 80 60 100 NZYBuffer U * NotI (MB153) DdeI C↓↓↓TNAG NZYBuffer U 25 25 25 100 NZYBuffer U No BstDEI, HpyF3I (MB236) MalI, BfuCI ♦♦♦, Bsp143I ♦♦♦, BstENII ♦♦♦, DpnI G(mA)↓↓↓TC NZYBuffer C 70 50 100 100 NZYBuffer U * (MB078) BstKTI ♦♦♦, BstMBI ♦♦♦, DpnII ♦♦♦, Kzo9I ♦♦♦, NdeII ♦♦♦ BfuCI, Bsp143I, BssMI, BstKTI, BstMBI, DpnII ↓↓↓GATC NZYBuffer C 80 80 100 100 NZYBuffer U * (MB233) Kzo9I, MboI, NdeII, Sau3AI EcoRI G↓↓↓AATTC NZYBuffer A 100 (120) (80) 100 NZYBuffer U * FunII (MB067) EcoRV GAT ↓↓↓ATC NZYBuffer A 100 0 25-50 100 NZYBuffer U * Eco32I (MB068) HindIII A↓↓↓AGCTT NZYBuffer B <20 100 (100) 100 NZYBuffer U * No None (MB070) HinfI G↓↓↓ANTC NZYBuffer C (90) (90) 100 100 NZYBuffer U * None (MB239) HpaI GTT ↓↓↓AAC NZYBuffer B 20 100 (80) 100 NZYBuffer U * KspAI (MB071) KpnI GGTAC ↓↓↓C NZYBuffer C 50 (80) 100 100 NZYBuffer U * No Acc65I ♦♦♦, Asp718I ♦♦♦ (MB072) BfuCI, Bsp143I, BstENII, BstMBI, DpnII, MboI ↓↓↓GATC NZYBuffer C 100 100 100 100 NZYBuffer U * (MB241) Kzo9II, NdeII, BstKTI ♦♦♦ MluI -

James Alan Tunaley
Structure and Activity Investigations of the Cell Fate Determinant, SpoIIE, from Bacillus subtilis. James Alan Tunaley Thesis Submitted for the Degree of Doctor of Philosophy University of York Department of Chemistry September 2013 Abstract For many years the Gram positive bacterium Bacillus subtilis has been a model organism for prokaryotic cell and molecular biology. The asymmetric cell division which B. subtilis undergoes during sporulation is a simple system by which to study the process of cell differentiation. Sporulation is governed by a series of genetic temporal and spatial controls. Gene regulation brought about by a series of σ factors and transcriptional regulators is coupled to key morphological stages or checkpoints. σF initiates the first step in a cascade of complex genetic control which eventually produces a resilient endospore. The activation of σF, the first compartment-specific sigma factor, in the forespore and its regulation through interaction between three proteins; SpoIIAA, SpoIIAB and SpoIIE, is of particular interest. SpoIIE, a protein phosphatase which binds to the asymmetric division septum, is a crucial factor in the selective activation of σF in the forespore. Of three putative domains in SpoIIE only the C-terminal PP2C phosphatase domain has been structurally characterised. The central domain, domain II, of SpoIIE has been assigned a role in interaction with the cell division machinery; however mutational studies have shown that, in addition, this domain is also responsible for the regulation of phosphatase activity. This work describes the isolation and characterisation of three new fragments of SpoIIE containing elements of the central cytoplasmic domain of SpoIIE. These include a fragment found to accurately represent the N-terminal solubility limit of domain II which shows a high degree of oligomeric character. -

Biology of DNA Restriction THOMAS A
MICROBIOLOGICAL REVIEWS, June 1993, p. 434-450 Vol. 57, No. 2 0146-0749/93/020434-17$02.00/0 Copyright © 1993, American Society for Microbiology Biology of DNA Restriction THOMAS A. BICKLE'* AND DETLEV H. KRUGER2 Department ofMicrobiology, Biozentrum, Basel University, Klingelbergstrasse 70, CH-4056 Basel, Switzerland, 1 and Institute of Virology, Charite School ofMedicine, Humboldt University, D-0-1040 Berlin, Gernany2 INTRODUCTION ........................................................................ 434 TYPE I R-M SYSTEMS ........................................................................ 434 Type I Systems Form Families of Related Enzymes ...................................................................435 Structure of hsd Genes ........................................................................ 435 Evolution of DNA Sequence Recognition by Recombination between hsdS Genes .........................***436 Mutations Affecting Modification Activity........................................................................ 437 TYPE II R-M SYSTEMS........................................................................ 437 Evolutionary Aspects ........................................................................ 437 Control of Expression of Type II RM Genes .....................................................438 Cytosine Can Be Methylated on Either C-5 Or NA: Consequences for Mutagenesis...............438 Type II Restriction Endonucleases That Require Two Recognition Sites for Cleavage.439 What Is the Function of Type IIS Enzymes.440 -

Curriculum Vitae SIR RICHARD JOHN ROBERTS ADDRESS PERSONAL
Curriculum Vitae SIR RICHARD JOHN ROBERTS ADDRESS New England Biolabs 240 County Road, Ipswich, MA 02138 USA Email: [email protected] Telephone: (978) 380-7405 / Fax: (978) 380-7406 PERSONAL Born on September 6, 1943, Derby, England EDUCATION 1962-1965 University of Sheffield, Sheffield, England B.Sc. in Chemistry 1966-1968 University of Sheffield, Sheffield, England Ph.D. in Organic Chemistry POSITIONS 2005- Chief Scientific Officer, New England Biolabs 1992-2005 Research Director, New England Biolabs 1986-92 Assistant Director for Research, Cold Spring Harbor Laboratory 1972-86 Senior Staff Investigator, Cold Spring Harbor Laboratory 1971-1972 Research Associate in Biochemistry, Harvard University 1969-1970 Research Fellow, Harvard University OUTSIDE ACTIVITIES 1974-1992 Consultant and Chairman of Scientific Advisory Board New England Biolabs 1977-1985 Scientific Advisory Board, Genex Corp. 1977-1987 Editorial Board: Nucleic Acids Research 1979-1984 Editorial Board: Journal of Biological Chemistry 1982-1989 Member: National Advisory Committee of GENBANK 1984-1986 Member: National Advisory Committee of BIONET 1985-1988 Panel member: NIH Study Section in Biochemistry. 1985-2002 Editorial Board: Bioinformatics (formerly CABIOS) 1987-1990 Chairman: National Advisory Committee of BIONET 1987-2009 Senior Executive Editor: Nucleic Acids Research 1990-1992 Panel member: NCI Cancer Centers Support Grant Review Committee 1993-1995 Panel member: NLM Study Section/Comp. Biol. 1994-2000 Scientific Advisory Board, Molecular Tool 1994- Patron of the Oxford International Biomedical Center 1996-1998 Visiting Professor, University of Bath, UK. 1996-2000 Chairman, NCI Board of Scientific Counselors 1996-1999 Scientific Advisory Board, Oxford Molecular Group 1997-2001 Editorial Board: Current Opinion Chem. Biol. -

Restriction Enzymes Are Molecular Scissors
Molecular Scissors Restriction enzymes are molecular scissors DR. ABUL HASAN SARDAR Assistant Professor and Head Department of Microbiology Sarsuna College RESTRICTION ENZYMES • A restriction enzyme (or restriction endonuclease) is an enzyme that cuts double- stranded or single stranded DNA at specific recognition nucleotide sequences known as restriction sites. Property of restriction enzymes • They that link adjacent nucleotides in DNA molecules. HOW RESTRICTION ENZYMES WORKS? • Restriction enzymes recognize a specific sequence of nucleotides, and produce a double-stranded cut in the DNA, these cuts are of two types: • BLUNT ENDS. • STICKY ENDS. Blunt end Sticky end BLUNT ENDS • These blunt ended fragments can be joined to any other DNA fragment with blunt ends. • Enzymes useful for certain types of DNA cloning experiments “STICKY ENDS” ARE USEFUL DNA fragments with complimentary sticky ends can be combined to create new molecules which allows the creation and manipulation of DNA sequences from different sources. • While recognition sequences vary widely , with lengths between 4 and 8 nucleotides, many of them are palindromic. PALINDROMES IN DNA SEQUENCES Genetic palindromes are similar to verbal palindromes. A palindromic sequence in DNA is one in which the 5’ to 3’ base pair sequence is identical on both strands (the 5’ and 3’ ends refers to the chemical structure of the DNA). PALINDROME SEQUENCES • The mirror like palindrome in which the same forward and backwards are on a single strand of DNA strand, as in GTAATG • The Inverted repeat palindromes is also a sequence that reads the same forward and backwards, but the forward and backward sequences are found in complementary DNA strands (GTATAC being complementary to CATATG) • Inverted repeat palindromes are more common and have greater biological importance than mirror- like palindromes.