UNIVERSITY of CALIFORNIA Los Angeles Avian
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RBMV011 a Diagnostic Tool to Identify Species of the Genus
A DIAGNOSTIC TOOL TO IDENTIFY SPECIES OF THE GENUS Isospora SCHNEIDER, 1881 (APICOMPLEXA: EIMERIIDAE) BASED ON SPORULATED OOCYSTS FROM THAUPIDAE FAMILY (AVES: PASSERIFORMES): A DICHOTOMOUS KEY* UMA FERRAMENTA DE DIAGNÓSTICO PARA IDENTIFICAR ESPÉCIES DE Isospora SCHNEIDER, 1881 (APICOMPLEXA: EIMERIIDAE) DA FAMÍLIA THAUPIDAE (AVES: PASSERIFORMES) COM BASE EM OOCISTOS ESPORULADOS: UMA CHAVE DICOTÔMICA Bruno Pereira Berto1, Hermes Ribeiro Luz2, Ildemar Ferreira3, Walter Flausino4 and Carlos Wilson Gomes Lopes5 ABSTRACT. Berto B.P., Luz, H.B., Ferreira I., Flausino W. & Lopes C.W.G. A diagnostic tool to identify species of the genus Isospora Schneider, 1881 (Apicomplexa: Eimeriidae) based on sporulated oocysts from Thaupidae family (Aves: Passeriformes): a dichotomous key [Uma ferramenta de diagnóstico para identificar espécies de Isospora Schneider, 1881 (Apicomplexa: Eimeriidae) da família Thaupidae (Aves: Passeriformes): uma chave dicotômica]. Revista Brasileira de Medicina Veterinária, 32(3):182-186, 2010. Laboratório de Coccídios e Coccidioses, Departamento de Parasitologia Animal, Instituto de Veterinária, Universidade Federal Rural do Rio de Janeiro, BR 465 km 7, Seropédica, RJ 23890-000, Brasil. E-mail: [email protected] The birds of the Thraupidae family, similar to other passerine birds can be infected by coccidia, and the genus Isospora can be considered the most relevant. Isospora thraupis, I. andesensis, I. irisidornisi, I. tiesangui, I. marambaiensis, I. sepetibensis, I. cadimi, I. navarroi, I. ramphoceli, I. sanhaci, I. sayacae and I. silvasouzai were described from thraupids of South American and considering that these birds are sympatric hosts of other birds that inhabit South, North and Central Americas, it is concluded that these parasites should be widely dispersed in the America Continent. -

Creación De Un Manual Interpretativo Para El Buen
CARRERA ADMINISTRACIÓN TURÍSTICA Y HOTELERA CREACIÓN DE UN MANUAL INTERPRETATIVO SOBRE EL BUEN AVISTAMIENTO DE AVES EN LA RESERVA YANACOCHA UBICADA EN LA PARROQUIA DE NONO CANTÓN QUITO PROVINCIA PICHINCHA CON EL PROPÓSITO DE DAR A CONOCER EL AVITURISMO Proyecto de investigación previo a la obtención de título de tecnólogo en Administración Turística y Hotelera Autora: Amanda Estefania Tituaña Espinosa Tutor: Ing. Ximena Almeida Quito, Diciembre 2018 i Declaratoria Declaro que la investigación es absolutamente original, autentica, personal, que se han citado las fuentes correspondientes y en su ejecución se respetaron las disposiciones legales que protegen los derechos de autor vigentes. Las ideas, doctrinas resultados y conclusiones a los que he llegado son de mi absoluta responsabilidad. Amanda Estefania Tituaña Espinosa CC 1750804161 CREACIÓN DE UN MANUAL INTERPRETATIVO SOBRE EL BUEN AVISTAMIENTO DE AVES EN LA RESERVA YANACOCHA UBICADA EN LA PARROQUIA DE NONO CANTON QUITO PROVINCIA PICHINCHA CON EL PROPOSITO DE DAR A CONOCER EL AVITURISMO ii Licencia De Uso No Comercial Yo, Amanda Estefania Tituaña Espinosa portadora de la cedula de ciudadanía asignada Con el No. 175080416-1 de conformidad con lo establecido en el Artículo 110 del Código de Economía Social de los Conocimientos, la Creación y la Innovación (INGENIOS) que dice: “En el caso de las obras creadas en centros educativos,universidades,escuelas politécnicas, institutos superiores tecnicos,tecnólogos, pedagógicos, de arte y los conservatorios superiores , e institutos públicos de investigación como resultado de su actividad académica o de investigación tales como trabajos de titulación, proyectos de investigación o innovación, articulo académico , u otros análogos , sin perjuicio de que pueda existir relación de dependencia , la titularidad de los derechos patrimoniales corresponderá a los autores . -

BIOTA COLOMBIANA ISSN Impreso 0124-5376 Volumen 20 · Número 1 · Enero-Junio De 2019 ISSN Digital 2539-200X DOI 10.21068/C001
BIOTA COLOMBIANA ISSN impreso 0124-5376 Volumen 20 · Número 1 · Enero-junio de 2019 ISSN digital 2539-200X DOI 10.21068/c001 Atropellamiento vial de fauna silvestre en la Troncal del Caribe Amaryllidaceae en Colombia Adiciones al inventario de copépodos de Colombia Nuevos registros de avispas en la región del Orinoco Herpetofauna de San José del Guaviare Escarabajos estercoleros en Aves en los páramos de Antioquia Oglán Alto, Ecuador y el complejo de Chingaza Biota Colombiana es una revista científica, periódica-semestral, Comité Directivo / Steering Committee que publica artículos originales y ensayos sobre la biodiversi- Brigitte L. G. Baptiste Instituto de Investigación de Recursos Biológicos dad de la región neotropical, con énfasis en Colombia y países Alexander von Humboldt vecinos, arbitrados mínimo por dos evaluadores externos. In- M. Gonzalo Andrade Instituto de Ciencias Naturales, Universidad Nacional de Colombia cluye temas relativos a botánica, zoología, ecología, biología, Francisco A. Arias Isaza Instituto de Investigaciones Marinas y Costeras limnología, conservación, manejo de recursos y uso de la bio- “José Benito Vives De Andréis” - Invemar diversidad. El envío de un manuscrito implica la declaración Charlotte Taylor Missouri Botanical Garden explícita por parte del (los) autor (es) de que este no ha sido previamente publicado, ni aceptado para su publicación en otra Editor / Editor revista u otro órgano de difusión científica. El proceso de arbi- Rodrigo Bernal Independiente traje tiene una duración mínima de tres a cuatro meses a partir Editor de artículos de datos / Data papers Editor de la recepción del artículo por parte de Biota Colombiana. To- Dairo Escobar Instituto de Investigación de Recursos Biológicos das las contribuciones son de la entera responsabilidad de sus Alexander von Humboldt autores y no del Instituto de Investigación de Recursos Bioló- Asistente editorial / Editorial assistant gicos Alexander von Humboldt, ni de la revista o sus editores. -

(Phthiraptera: Amblycera and Ischnocera) on Birds of Peru
Arxius de Miscel·lània Zoològica, 19 (2021): 7–52 ISSN:Minaya 1698– et0476 al. Checklist of chewing lice (Phthiraptera: Amblycera and Ischnocera) on birds of Peru D. Minaya, F. Príncipe, J. Iannacone Minaya, D., Príncipe, F., Iannacone, J., 2021. Checklist of chewing lice (Phthiraptera: Am- blycera and Ischnocera) on the birds of Peru. Arxius de Miscel·lània Zoològica, 19: 7–52, Doi: https://doi.org/10.32800/amz.2021.19.0007 Abstract Checklist of chewing lice (Phthiraptera: Amblycera and Ischnocera) on birds of Peru. Peru is one of the countries with the highest diversity of birds worldwide, having about 1,876 species in its territory. However, studies focused on chewing lice (Phthiraptera) have been carried out on only a minority of bird species. The available data are distributed in 87 publications in the national and international literature. In this checklist we summarize all the records to date of chewing lice on wild and domestic birds in Peru. Among the 301 species of birds studied, 266 species of chewing lice were recorded. The localities with the highest records were the Departments of Cusco, Junín, Lima and Madre de Dios. No records of birds pa- rasitized by these lice have been found in seven departments of Peru. Studies related to lice have only been reported in 16 % of bird species in the country, indicating that research concerning chewing lice has not yet been performed for the the majority of birds in Peru. Data published through GBIF (Doi: 10.15470/u1jtiu) Key words: Avifauna, Ectoparasites, Lice, Parasitology, Phthiraptera Resumen Lista de verificación de piojos masticadores (Phthiraptera: Amblycera e Ischnocera) de las aves de Perú. -

Ufrrj Instituto De Veterinária Curso De Pós-Graduação
UFRRJ INSTITUTO DE VETERINÁRIA CURSO DE PÓS-GRADUAÇÃO EM CIÊNCIAS VETERINÁRIAS DISSERTAÇÃO ESPÉCIES DE COCCÍDIOS EM THRAUPIDAE (AVES: PASSERIFORMES) DO PARQUE NACIONAL DO ITATIAIA, RJ: MORFOLOGIA E TAXONOMIA MARIANA BORGES RODRIGUES 2016 UNIVERSIDADE FEDERAL RURAL DO RIO DE JANEIRO INSTITUTO DE VETERINÁRIA CURSO DE PÓS-GRADUAÇÃO EM CIÊNCIAS VETERINÁRIAS ESPÉCIES DE COCCÍDIOS EM THRAUPIDAE (AVES: PASSERIFORMES) DO PARQUE NACIONAL DO ITATIAIA, RJ: MORFOLOGIA E TAXONOMIA MARIANA BORGES RODRIGUES Sob a orientação do Professor Dr. Bruno Pereira Berto Dissertação submetida como requisito parcial para obtenção do grau de Mestre em Ciências, no Curso de Pós-Graduação em Ciências Veterinárias. Seropédica, RJ Fevereiro de 2016 UNIVERSIDADE FEDERAL RURAL DO RIO DE JANEIRO INSTITUTO DE VETERINÁRIA CURSO DE PÓS-GRADUAÇÃO EM CIÊNCIAS VETERINÁRIAS MARIANA BORGES RODRIGUES Dissertação submetida como requisito parcial para obtenção do grau de Mestre em Ciências, no Curso de Pós-Graduação em Ciências Veterinárias. DISSERTAÇÃO APROVADA EM 19/02/2016 DEDICATÓRIA Ao Senhor meu Deus pai, fiel em todas as suas promessas, que guia e protege os meus caminhos. Aos meus queridos pais Jurandir de Jesus Rodrigues e Célia Borges Rodrigues, pelas orações, apoio e incentivo. A Juliana Borges Rodrigues e Rodrigues Reis e Davi Daniel Magalhães Reis pelas palavras amigas e companheirismo. Aos familiares que me apoiaram e acreditaram em mim. AGRADECIMENTOS Agradeço a todos os colegas que contribuíram direta ou indiretamente na obtenção dos dados de pesquisa e processamentos laboratoriais que permitiram a finalização deste trabalho. Agradeço em especial ao Prof. Dr. Bruno Pereira Berto, do Laboratório de Coccídios e Coccidioses (LCC), Departamento de Biologia Animal (DBA) do Instituto de Ciências Biólogicas e da Saúde (ICBS), Universidade Federal Rural do Rio de Janeiro (UFRRJ), Campus Seropédica, RJ pela amizade, incentivo, atenção, direcionamento e esforços para que esta pesquisa alcançasse os seus objetivos. -

2. Birds of South America
TRAFFIC Bird’s-eye view: REPORT Lessons from 50 years of bird trade regulation & conservation in Amazon countries DECEMBER 2018 Bernardo Ortiz-von Halle About the author and this study: Bernardo Ortiz-von Halle, a biologist and TRAFFIC REPORT zoologist from the Universidad del Valle, Cali, Colombia, has more than 30 years of experience in numerous aspects of conservation and its links to development. His decades of work for IUCN - International Union for Conservation of Nature and TRAFFIC TRAFFIC, the wildlife trade monitoring in South America have allowed him to network, is a leading non-governmental organization working globally on trade acquire a unique outlook on the mechanisms, in wild animals and plants in the context institutions, stakeholders and challenges facing of both biodiversity conservation and the conservation and sustainable use of species sustainable development. and ecosystems. Developing a critical perspective The views of the authors expressed in this of what works and what doesn’t to achieve lasting conservation goals, publication do not necessarily reflect those Bernardo has put this expertise within an historic framework to interpret of TRAFFIC, WWF, or IUCN. the outcomes of different wildlife policies and actions in South America, Reproduction of material appearing in offering guidance towards solutions that require new ways of looking at this report requires written permission wildlife trade-related problems. Always framing analysis and interpretation from the publisher. in the midst of the socioeconomic and political frameworks of each South The designations of geographical entities in American country and in the region as a whole, this work puts forward this publication, and the presentation of the conclusions and possible solutions to bird trade-related issues that are material, do not imply the expression of any linked to global dynamics, especially those related to wildlife trade. -
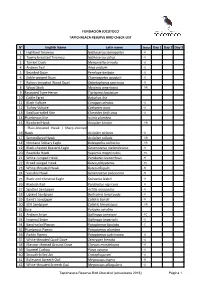
N° English Name Latin Name Status Day 1 Day 2
FUNDACIÓN JOCOTOCO TAPICHALACA RESERVE BIRD CHECK-LIST N° English Name Latin name Status Day 1 Day 2 Day 3 1 Highland Tinamou Nothocercus bonapartei - R 2 Tawny-breasted Tinamou Nothocercus julius - U 3 Torrent Duck Merganetta armata - U 4 Andean Teal Anas andium - U 5 Bearded Guan Penelope barbata - U 6 Sickle-winged Guan Chamaepetes goudotii - U 7 Rufous-breasted Wood Quail Odontophorus speciosus - R 8 Wood Stork Mycteria americana - VR 9 Fasciated Tiger-Heron Tigrisoma fasciatum 10 Cattle Egret Bubulcus ibis 11 Black Vulture Coragyps atratus - U 12 Turkey Vulture Cathartes aura - U 13 Swallow-tailed Kite Elanoides forficatus - U 14 Plumbeous Kite Ictinia plumbea 15 Bicolored Hawk Accipiter bicolor - VR Plain-breasted Hawk / Sharp-shinned 16 Hawk Accipiter striatus - U 17 Semicollared Hawk Accipiter collaris - VR 18 Montane Solitary Eagle Buteogallus solitarius - VR 19 Black-chested Buzzard-Eagle Geranoaetus melanoleucus - R 20 Roadside Hawk Rupornis magnirostris - FC 21 White-rumped Hawk Parabuteo leucorrhous - R 22 Broad-winged Hawk Buteo platypterus - FC 23 White-throated Hawk Buteo albigula - R 24 Variable Hawk Geranoaetus polyosoma - R 25 Black-and-chestnut Eagle Spizaetus isidori - R 26 Blackish Rail Pardirallus nigricans - R 27 Spotted Sandpiper Actitis macularius - R 28 Upland Sandpiper Bartramia longicauda - R 29 Baird's Sandpiper Calidris bairdii - R 30 Stilt Sandpiper Calidris himantopus - VR 31 Sora Porzana carolina 32 Andean Snipe Gallinago jamesoni - FC 33 Imperial Snipe Gallinago imperialis - U 34 Band-tailed Pigeon Patagioenas -

The Birds of Abra Patricia and the Upper Río Mayo, San Martín, North
T h e birds of Ab r a Patricia and the upper río Mayo, San Martín, north Peru Jon Hornbuckle Cotinga 12 (1999): 11– 28 En 1998 se llevó a cabo un inventario ornitológico en un bosque al este de Abra Patricia, Departamento San Martín, norte de Perú, en el cual se registraron 317 especies de aves. Junto con los registros previamente publicados y observaciones recientes realizadas por visitantes al área, el número de especies asciende a por lo menos 420. De éstas, 23 están clasificadas como amenazadas globalmente3, incluyendo Xenoglaux loweryi y Grallaricula ochraceifrons, ambas prácticamente desconocidas. Además, se registraron siete especies de distribución restringida. A pesar de que el ‘Bosque de Protección del Alto Mayo’ protege teóricamente 182 000 ha, la tala del bosque es una actividad frecuente y al parecer no existen medidas reales de control. En la actualidad se están realizando esfuerzos para conservar esta importante área. Introduction ochraceifrons10,15. However, ornithological surveys of In northern Peru, the forest east of the Abra Patricia this area have been confined to three Louisiana pass, dpto. San Martin (see Appendix 3 for State University Museum of Zoology (LSUMZ) coordinates) is of particular interest to expeditions, totalling six weeks: in 1976, 1977 and ornithologists as it is the type-locality for the near- 19835,15,18. Since that period the region has been too mythical Long-whiskered Owlet Xenoglaux loweryi dangerous to visit, until the recent cessation of and Ochre-fronted Antpitta Grallaricula guerilla activities. 11 Cotinga 12 The birds of Abra Patricia and the upper río Mayo, San M artín, north Peru The area is located at the northern end of the The habitat at the LSUMZ study sites has been Cordillera Oriental, the easternmost range of the described in some detail5,15,17 but can be summarised north Peruvian Andes, sloping eastward to the Rio at the lower elevations as subtropical forest of tall Mayo. -

Proceedings of the Indiana Academy of Science (1988) Volume 98 P
Proceedings of the Indiana Academy of Science (1988) Volume 98 p. 581-593. SKELETONS AND THE GENERA OF TANAGERS J. Dan Webster Hanover College Hanover, Indiana 47243 ABSTRACT: There are 248 species of tanagers ( Aves, Pas- seriformes, Thraupinae) arranged in 61 genera and known chiefly from their skins. The author studied 434 skeletons of 191 species of 57 genera, describing the shape or measuring 36 characters on each. Twenty-three characters proved useful in generic discrimination. Presence or absence of a free lac- rimal; presence or absence of a manubrium-sternum bridge; shape and size of the interpalatine process; and the tibiotar- sus/ulna length ratio were the best discriminants. The phy- logeny of the tanagers remains unclear. More specimens must be studied. INTRODUCTION The 248 species of tanagers (Passeriformes, Emberizidae, Thraupinae) con- stitute a large, varied, colorful subfamily of neotropical birds. The last systematist to list all the known genera and species with their characters was Sclater (1886). Ridgway (1902) differentiated all the genera and those species found in North and Central American and the West Indies. Storer discussed the classification (1969) and listed all the forms (1970). Other important contributions were by Wetmore (1914), who described the peculiar stomach ofEuphonia, and by Beecher (1951), who stated, mainly on the basis of jaw muscles, that several genera of honeycreepers belonged with the tanagers. Lucas (1894) and Bock (1985) differ- entiated genera of honeycreepers on the basis of tongue structure. Sibley (1970) studied the egg white proteins of 7 genera. Raikow (1978, 1985) devised a clas- sification based mainly on the appendicular myology of 13 genera plus 4 borderline genera. -

New Isospora and Host Species in Brazilian Passerines
Brazilian Journal of Poultry Science Revista Brasileira de Ciência Avícola ISSN 1516-635X 2020 / v.22 / n.1 / 001-014 New Isospora and Host Species in Brazilian Passerines http://dx.doi.org/10.1590/1806-9061-2019-1070 Original Article Author(s) ABSTRACT Barreto CI https://orcid.org/0000-0002-6890-3746 Normal passerines (n=216) were evaluated for oocysts of Isospora Vilela DARI https://orcid.org/0000-0001-5707-3233 in feces at the Triage Center for Wild Animals (CETAS, IBAMA, Belo Houri BFII https://orcid.org/0000-0002-6659-3887 Horizonte; August 21 to September 21, 2012). The positive samples III Lara LB https://orcid.org/0000-0002-3995-0949 with oocysts represented 13.0% of Cardinalidae (n=23), 11.2% Torres ACDIV https://orcid.org/0000-0002-7199-6517 Silva ASGIV https://orcid.org/0000-0002 3195 9586 of Emberizidae (n=107), 50% of Icteridae (n=10) and 60.3% of Castro Filho RPLIV https://orcid.org/0000-0002-9300-7513 Thraupidae (n=68). The probability of fecal oocysts attributable to Costa CSIV https://orcid.org/0000-0003-0701-1733 the host in Thraupidae is higher than in Cardinalidae, Emberizidae, Martins NRSIV https://orcid.org/0000-0001-8925-2228 Fringillidae and Turdidae, but similar to Icteridae. No oocysts were I CETAS, Instituto Brasileiro de Meio Ambiente e found in Fringillidae and Turdidae. Within Thraupidae, Isospora was Recursos Naturais Renováveis, IBAMA, Avenida for the first time described in Paroaria dominicana and Schistochlamys do Contorno, Nº 8.121, Cidade Jardim, CEP 30.110-120, Belo Horizonte, MG, Brazil. ruficapillus and within Icteridae, in Gnorimopsar chopi. -

Bulletin of the British Ornithologists' Club
Bulletin of the British Ornithologists’ Club Volume 133 No. 4 December 2013 FORTHCOMING MEETINGS See also BOC website: http://www.boc-online.org BOC MEETINGS are open to all, not just BOC members, and are free. Evening meetings are in an upstairs room at The Barley Mow, 104, Horseferry Road, Westminster, London SW1P 2EE. The nearest Tube stations are Victoria and St James’s Park; and the 507 bus, which runs from Victoria to Waterloo, stops nearby. For maps, see http://www.markettaverns.co.uk/the_barley_mow.html or ask the Chairman for directions. The cash bar opens at 6.00 pm and those who wish to eat after the meeting can place an order. The talk will start at 6.30 pm and, with questions, will last c.1 hour. It would be very helpful if those intending to come can notify the Chairman no later than the day before the meeting. 25 February 2014—6.30 pm—Richard Porter—Birds of Socotra: populations and distribution Abstract: The Socotra archipelago lies in the Arabian Sea, c.350 km south of the Yemen mainland. Ecologically heavily influenced by the dry south-west monsoon that batters the islands in May–September, it boasts high endemism in plants, reptiles, insects and birds, which I will summarise. Whilst its avifauna is species-poor, with just 42 regular breeders, 11 are endemic, making it the richest area of avian endemism in the Middle East (along with the highlands of south-west Arabia). From 1999 to 2011, I have been engaged in mapping the distribution and determining the populations of the breeding species and I will present some of my findings. -

Thraupinae Species Tree
Thraupinae I: Catamblyrhynchini through Cissopini Plushcap, Catamblyrhynchus diadema Catamblyrhynchini Mourning Sierra-Finch, Rhopospina fruticeti Blue Finch, Rhopospina caerulescens Band-tailed Sierra Finch, Rhopospina alaudina Carbonated Sierra Finch, Rhopospina carbonaria Porphyrospizini Great Inca-Finch, Incaspiza pulchra Rufous-backed Inca-Finch, Incaspiza personata Gray-winged Inca-Finch, Incaspiza ortizi Buff-bridled Inca-Finch, Incaspiza laeta Little Inca-Finch, Incaspiza watkinsi Yellow-shouldered Grosbeak, Parkerthraustes humeralis Orchesticini Brown Tanager, Orchesticus abeillei Vermilion Tanager, Calochaetes coccineus Purplish-mantled Tanager, Iridosornis porphyrocephalus Yellow-throated Tanager, Iridosornis analis Golden-collared Tanager, Iridosornis jelskii Golden-crowned Tanager, Iridosornis rufivertex Yellow-scarfed Tanager, Iridosornis reinhardti Pipraeideini Fawn-breasted Tanager, Pipraeidea melanonota Blue-and-yellow Tanager, Pipraeidea bonariensis Rufous-bellied Mountain-Tanager, Pseudosaltator rufiventris Buff-breasted Mountain-Tanager, Dubusia taeniata Chestnut-bellied Mountain-Tanager, Dubusia castaneoventris Hooded Mountain-Tanager, Buthraupis montana Blue-capped Tanager, Sporathraupis cyanocephala Masked Mountain-Tanager, Tephrophilus wetmorei Grass-green Tanager, Chlorornis riefferii Black-chested Mountain-Tanager, Cnemathraupis eximia Golden-backed Mountain-Tanager, Cnemathraupis aureodorsalis Blue-winged Mountain-Tanager, Compsocoma somptuosa Black-chinned Mountain-Tanager, Compsocoma notabilis Black-cheeked (Santa