A Genus-Level Classification of the Family Thraupidae (Class Aves: Order Passeriformes)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cross-Temporal Analysis of Genetic Diversity in the Endangered Medium Tree Finch (Camarhynchus Pauper) and Closely Related Darwin’S Finches
Cross-Temporal Analysis of Genetic Diversity in the Endangered Medium Tree Finch (Camarhynchus pauper) and Closely Related Darwin’s Finches By Colleen Metzger B.S., Juniata College, 2009 A thesis submitted to the Graduate School of the University of Cincinnati Department of Biological Sciences In partial fulfillment of the requirements For the degree of Master of Science Committee Chair: Kenneth Petren, Ph.D. November 2012 Abstract Natural history collections can provide a direct view of past genotypes, which allows greater insight into evolutionary processes that are relevant for conservation and management. However, few studies have used broad surveys of multilocus genotypes from the past to address the wide range of processes that can affect conservation planning of a species today. Therefore, we assessed the history and status of the critically endangered medium tree finch, Camarhynchus pauper, an endemic finch of the Galápagos Islands. Using ancient DNA techniques, we quantified cross-temporal genetic change for 16 microsatellite loci in this species and its relatives. We tested the hypothesis that C. pauper has undergone a recent reduction in population size and loss of genetic diversity, and evaluated the hypothesis that C. pauper is genetically distinct from its two closest relatives, C. parvulus and C. psittacula. We assessed whether decline in C. pauper has led to increased hybridization with other species and evaluated a long-standing hypothesis of its origin from C. psittacula on another island using genetic distances, assignment tests, and migration analyses. Genetic diversity declined significantly in C. pauper over time, and several other tree finch populations showed similar losses of genetic diversity. -

Wings Without Borders Alas Sin Fronteras IV North American Ornithological Conference IV Congreso Norteamericano De Ornitología
Wings Without Borders Alas Sin Fronteras IV North American Ornithological Conference IV Congreso Norteamericano de Ornitología October 3-7, 2006 · 3-7 Octubre 2006 Veracruz, México CONFERENCE PROGRAM PROGRAMA DEL CONGRESO IV NAOC is organized jointly by the American Ornithologists’ Union, Association of Field Ornithologists, Sección Mexicana de Consejo Internacional para la Preservación de las Aves, A. C., Cooper Ornithological Society, Raptor Research Foundation, Society of Canadian Ornithologists / Société des Ornithologistes du Canada, Waterbird Society, and Wilson Ornithological Society 4to. Congreso Norteamericano de Ornitología - Alas Sin Fronteras Programa del Congreso Table of Contents IV NAOC Conference Committees ......................................................................................................................................................................................2 Local Hosts ...........................................................................................................................................................................................................................2 Conference Sponsors .............................................................................................................................................................................................................3 Other Sponsors ....................................................................................................................................................................................................................3 -
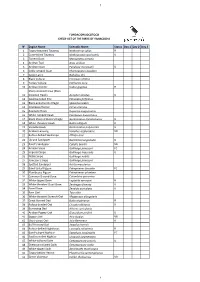
N° English Name Scientific Name Status Day 1
1 FUNDACIÓN JOCOTOCO CHECK-LIST OF THE BIRDS OF YANACOCHA N° English Name Scientific Name Status Day 1 Day 2 Day 3 1 Tawny-breasted Tinamou Nothocercus julius R 2 Curve-billed Tinamou Nothoprocta curvirostris U 3 Torrent Duck Merganetta armata 4 Andean Teal Anas andium 5 Andean Guan Penelope montagnii U 6 Sickle-winged Guan Chamaepetes goudotii 7 Cattle Egret Bubulcus ibis 8 Black Vulture Coragyps atratus 9 Turkey Vulture Cathartes aura 10 Andean Condor Vultur gryphus R Sharp-shinned Hawk (Plain- 11 breasted Hawk) Accipiter striatus U 12 Swallow-tailed Kite Elanoides forficatus 13 Black-and-chestnut Eagle Spizaetus isidori 14 Cinereous Harrier Circus cinereus 15 Roadside Hawk Rupornis magnirostris 16 White-rumped Hawk Parabuteo leucorrhous 17 Black-chested Buzzard-Eagle Geranoaetus melanoleucus U 18 White-throated Hawk Buteo albigula R 19 Variable Hawk Geranoaetus polyosoma U 20 Andean Lapwing Vanellus resplendens VR 21 Rufous-bellied Seedsnipe Attagis gayi 22 Upland Sandpiper Bartramia longicauda R 23 Baird's Sandpiper Calidris bairdii VR 24 Andean Snipe Gallinago jamesoni FC 25 Imperial Snipe Gallinago imperialis U 26 Noble Snipe Gallinago nobilis 27 Jameson's Snipe Gallinago jamesoni 28 Spotted Sandpiper Actitis macularius 29 Band-tailed Pigeon Patagoienas fasciata FC 30 Plumbeous Pigeon Patagioenas plumbea 31 Common Ground-Dove Columbina passerina 32 White-tipped Dove Leptotila verreauxi R 33 White-throated Quail-Dove Zentrygon frenata U 34 Eared Dove Zenaida auriculata U 35 Barn Owl Tyto alba 36 White-throated Screech-Owl Megascops -

Ornithological Surveys in Serranía De Los Churumbelos, Southern Colombia
Ornithological surveys in Serranía de los Churumbelos, southern Colombia Paul G. W . Salaman, Thomas M. Donegan and Andrés M. Cuervo Cotinga 12 (1999): 29– 39 En el marco de dos expediciones biológicos y Anglo-Colombian conservation expeditions — ‘Co conservacionistas anglo-colombianas multi-taxa, s lombia ‘98’ and the ‘Colombian EBA Project’. Seven llevaron a cabo relevamientos de aves en lo Serranía study sites were investigated using non-systematic de los Churumbelos, Cauca, en julio-agosto 1988, y observations and standardised mist-netting tech julio 1999. Se estudiaron siete sitios enter en 350 y niques by the three authors, with Dan Davison and 2500 m, con 421 especes registrados. Presentamos Liliana Dávalos in 1998. Each study site was situ un resumen de los especes raros para cada sitio, ated along an altitudinal transect at c. 300- incluyendo los nuevos registros de distribución más m elevational steps, from 350–2500 m on the Ama significativos. Los resultados estabilicen firme lo zonian slope of the Serranía. Our principal aim was prioridad conservacionista de lo Serranía de los to allow comparisons to be made between sites and Churumbelos, y aluco nos encontramos trabajando with other biological groups (mammals, herptiles, junto a los autoridades ambientales locales con insects and plants), and, incorporating geographi cuiras a lo protección del marcizo. cal and anthropological information, to produce a conservation assessment of the region (full results M e th o d s in Salaman et al.4). A sizeable part of eastern During 14 July–17 August 1998 and 3–22 July 1999, Cauca — the Bota Caucana — including the 80-km- ornithological surveys were undertaken in Serranía long Serranía de los Churumbelos had never been de los Churumbelos, Department of Cauca, by two subject to faunal surveys. -

Dacninae Species Tree, Part I
Dacninae I: Nemosiini, Conirostrini, & Diglossini Hooded Tanager, Nemosia pileata Cherry-throated Tanager, Nemosia rourei Nemosiini Blue-backed Tanager, Cyanicterus cyanicterus White-capped Tanager, Sericossypha albocristata Scarlet-throated Tanager, Sericossypha loricata Bicolored Conebill, Conirostrum bicolor Pearly-breasted Conebill, Conirostrum margaritae Chestnut-vented Conebill, Conirostrum speciosum Conirostrini White-eared Conebill, Conirostrum leucogenys Capped Conebill, Conirostrum albifrons Giant Conebill, Conirostrum binghami Blue-backed Conebill, Conirostrum sitticolor White-browed Conebill, Conirostrum ferrugineiventre Tamarugo Conebill, Conirostrum tamarugense Rufous-browed Conebill, Conirostrum rufum Cinereous Conebill, Conirostrum cinereum Stripe-tailed Yellow-Finch, Pseudochloris citrina Gray-hooded Sierra Finch, Phrygilus gayi Patagonian Sierra Finch, Phrygilus patagonicus Peruvian Sierra Finch, Phrygilus punensis Black-hooded Sierra Finch, Phrygilus atriceps Gough Finch, Rowettia goughensis White-bridled Finch, Melanodera melanodera Yellow-bridled Finch, Melanodera xanthogramma Inaccessible Island Finch, Nesospiza acunhae Nightingale Island Finch, Nesospiza questi Wilkins’s Finch, Nesospiza wilkinsi Saffron Finch, Sicalis flaveola Grassland Yellow-Finch, Sicalis luteola Orange-fronted Yellow-Finch, Sicalis columbiana Sulphur-throated Finch, Sicalis taczanowskii Bright-rumped Yellow-Finch, Sicalis uropigyalis Citron-headed Yellow-Finch, Sicalis luteocephala Patagonian Yellow-Finch, Sicalis lebruni Greenish Yellow-Finch, -

Reference File
References added since publication of 2007 CRC Handbook of Avian Body Masses Abadie, K. B., J. Pérez Z., and M. Valverde. 2006. Primer reporte de colonias del Martín Peruano Progne murphyi. Cotinga 24:99-101. Ackerman, J. T., J. Y. Takekawa, J. D. Bluso, J. L. Yee, and C. A. Eagles-Smith. 2008. Gender identification of Caspian Terns using external morphology and discriminant function analysis. Wilson Journal of Ornithology 120:378-383. Alarcos, S., C. de la Cruz, E. Solís, J. Valencia, and M. J. García-Baquero. 2007. Sex determination of Iberian Azure-winged Magpies Cyanopica cyanus cooki by discriminant analysis of external measurements. Ringing & Migration 23:211-216. Albayrak, T., A. Besnard, and A. Erdoğan. 2011. Morphometric variation and population relationships of Krüeper’s Nuthatch (Sitta krueperi) in Turkey. Wilson Journal of Ornithology 123:734-740. Aleixo, A., C. E. B. Portes, A. Whittaker, J. D. Weckstein, L. Pedreira Gonzaga, K. J. Zimmer, C. C. Ribas, and J. M. Bates. 2013. Molecular systematics and taxonomic revision of the Curve-billed Scythebill complex (Campylorhamphus procurvoides: Dendrocolaptidae), with description of a new species from western Amazonian Brazil. Pp. 253-257, In: del Hoyo, J., A Elliott, J. Sargatal, and D.A. Christie (eds). Handbook of the birds of the world. Special volume: new species and global index. Lynx Edicions, Barcelona, Spain. Volume 1. Alfano, A. 2014. Pygmy Nightjar (Nyctopolus hirundinaeus). Neotropical Birds Online (T.S. Schulenberg, ed.). Cornell Laboratory of Ornithology, Ithaca, NY. Alvarenga, H. M. F., E. Höfling, and L. F. Silveira. 2002. Notharchus swainsoni (Gray, 1846) é uma espécie válida. -

Download Download
Journal of Caribbean Ornithology RESEARCH ARTICLE Vol. 33:1–14. 2020 Composition of bird community in Portachuelo Pass (Henri Pittier National Park, Venezuela) Cristina Sainz-Borgo Jhonathan Miranda Miguel Lentino Photo: Pedro Arturo Amaro Journal of Caribbean Ornithology jco.birdscaribbean.org ISSN 1544-4953 RESEARCH ARTICLE Vol. 33:1–14. 2020 birdscaribbean.org Composition of bird community in Portachuelo Pass (Henri Pittier National Park, Venezuela) Cristina Sainz-Borgo1, Jhonathan Miranda2, and Miguel Lentino3 Abstract The purpose of this study was to describe the composition of the bird community in Portachuelo Pass, located in Henri Pittier National Park, Venezuela. Portachuelo Pass is an important route for migratory birds between northern South America and the Southern Cone. During 11 months of sampling between 2010 and 2012, we captured 1,460 birds belonging to 125 identified species, 29 families, and 9 orders. The families with the highest relative abundance and species richness were Trochilidae and Thraupidae and the most common species were the Violet-chested Hummingbird (Sternoclyta cyanopectus), Olive-striped Flycatcher (Mionectes olivaceus), Plain-brown Woodcreeper (Dendrocincla fuliginosa), Orange-bellied Euphonia (Euphonia xanthogaster), Violet-fronted Brilliant (Heliodoxa leadbeateri), Vaux’s Swift (Chaetura vauxi), Red-eared Parakeet (Pyrrhura hoematotis), Golden-tailed Sapphire (Chrysuronia oenone), Black-hooded Thrush (Turdus olivater), and Gray-rumped Swift (Chaetura cinereiventris). These species represented 52.4% of total captures and 8.0% of identified species. We captured 5 endemic species and 8 migratory species. The months of greatest relative abundance and species richness were June and July 2010 and January 2011. Birds captured belonged to the following feeding guilds: insectivorous, nectarivorous-insectivorous, frugivorous, frugivorous-insectivorous, granivorous, frugivorous-folivorous, omnivorous, carnivorous, and frugivorous-graniv- orous. -

Costa Rica: the Introtour | July 2017
Tropical Birding Trip Report Costa Rica: The Introtour | July 2017 A Tropical Birding SET DEPARTURE tour Costa Rica: The Introtour July 15 – 25, 2017 Tour Leader: Scott Olmstead INTRODUCTION This year’s July departure of the Costa Rica Introtour had great luck with many of the most spectacular, emblematic birds of Central America like Resplendent Quetzal (photo right), Three-wattled Bellbird, Great Green and Scarlet Macaws, and Keel-billed Toucan, as well as some excellent rarities like Black Hawk- Eagle, Ochraceous Pewee and Azure-hooded Jay. We enjoyed great weather for birding, with almost no morning rain throughout the trip, and just a few delightful afternoon and evening showers. Comfortable accommodations, iconic landscapes, abundant, delicious meals, and our charismatic driver Luís enhanced our time in the field. Our group, made up of a mix of first- timers to the tropics and more seasoned tropical birders, got along wonderfully, with some spying their first-ever toucans, motmots, puffbirds, etc. on this trip, and others ticking off regional endemics and hard-to-get species. We were fortunate to have several high-quality mammal sightings, including three monkey species, Derby’s Wooly Opossum, Northern Tamandua, and Tayra. Then there were many www.tropicalbirding.com +1-409-515-9110 [email protected] Page Tropical Birding Trip Report Costa Rica: The Introtour | July 2017 superb reptiles and amphibians, among them Emerald Basilisk, Helmeted Iguana, Green-and- black and Strawberry Poison Frogs, and Red-eyed Leaf Frog. And on a daily basis we saw many other fantastic and odd tropical treasures like glorious Blue Morpho butterflies, enormous tree ferns, and giant stick insects! TOP FIVE BIRDS OF THE TOUR (as voted by the group) 1. -

UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS Departamento De Zoología Y Antropología Física
UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS Departamento de Zoología y Antropología Física TESIS DOCTORAL Diversidad y especificidad de simbiontes en aves neotropicales Diversity and host specificity of symbionts in neotropical birds MEMORIA PARA OPTAR AL GRADO DE DOCTOR PRESENTADA POR Michaël André Jean Moens Directores Javier Pérez Tris Laura Benítez Rico Madrid, 2017 © Michaël André Jean Moens, 2016 Diversidad y Especificidad de Simbiontes en Aves Neotropicales (Diversity and Host Specificity of Symbionts in Neotropical birds.) Tesis doctoral de: Michaël André Jean Moens Directores : Javier Pérez Tris Laura Benítez Rico Madrid, 2016 © Michaël André Jean Moens, 2016 Cover Front Chestnut-breasted Coronet (Boissoneaua matthewsii) Taken at the San Isidro Reserve, Ecuador. Copyright © Jaime Culebras Cover Back Royal Flycatcher (Onychorhynchus coronatus) Taken at the Nouragues Reserve, French Guiana. Copyright © Borja Milá UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS DEPARTAMENTO DE ZOOLOGÍA Y ANTROPOLOGÍA FÍSICA Diversidad y Especificidad de Simbiontes en Aves Neotropicales (Diversity and Host Specificity of Symbionts in Neotropical birds.) Tesis doctoral de: Michaël André Jean Moens Directores: Javier Pérez Tris Laura Benítez Rico Madrid, 2016 © Michaël André Jean Moens, 2016 UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS DEPARTAMENTO DE ZOOLOGÍA Y ANTROPOLOGÍA FÍSICA Diversidad y Especificidad de Simbiontes en Aves Neotropicales (Diversity and Host Specificity of Symbionts -

21 Sep 2018 Lists of Victims and Hosts of the Parasitic
version: 21 Sep 2018 Lists of victims and hosts of the parasitic cowbirds (Molothrus). Peter E. Lowther, Field Museum Brood parasitism is an awkward term to describe an interaction between two species in which, as in predator-prey relationships, one species gains at the expense of the other. Brood parasites "prey" upon parental care. Victimized species usually have reduced breeding success, partly because of the additional cost of caring for alien eggs and young, and partly because of the behavior of brood parasites (both adults and young) which may directly and adversely affect the survival of the victim's own eggs or young. About 1% of all bird species, among 7 families, are brood parasites. The 5 species of brood parasitic “cowbirds” are currently all treated as members of the genus Molothrus. Host selection is an active process. Not all species co-occurring with brood parasites are equally likely to be selected nor are they of equal quality as hosts. Rather, to varying degrees, brood parasites are specialized for certain categories of hosts. Brood parasites may rely on a single host species to rear their young or may distribute their eggs among many species, seemingly without regard to any characteristics of potential hosts. Lists of species are not the best means to describe interactions between a brood parasitic species and its hosts. Such lists do not necessarily reflect the taxonomy used by the brood parasites themselves nor do they accurately reflect the complex interactions within bird communities (see Ortega 1998: 183-184). Host lists do, however, offer some insight into the process of host selection and do emphasize the wide variety of features than can impact on host selection. -

A Nest of Orange-Throated Tanager Wetmorethraupis Sterrhopteron
Cotinga 36 Short Communications in the region is highly fragmented, not yet relocated the nest itself and the forest surrounding the and did not observe any activity nest was c.10 ha. therein. However, they observed While observing three adult- no adults carrying food or nest plumaged Wetmorethraupis material. As the terrain is very moving through the canopy of steep, the group was only detected the forest fragment from a dirt >300 m from the nest on several road, we noted that they were occasions (moving to or from the repeatedly visiting a particular nest). Therefore, based on the ease site within a palm tree and further with which this species is detected observations using binoculars and by its loud vocalisations, we telescope revealed that at least two consider that most of the periods birds were constructing a nest. of absence were spent in forest The nest was c.10 m above fragments other than that of the the steeply sloping ground, nest, and that the adults may have in the uppermost fronds of a moved 1 km or more during these walking palm Socratea exhorrhiza forays. Based on the extensive A nest of Orange-throated (identified as probably this experience of HFG with other Tanager Wetmorethraupis species by A. Henderson, New (albeit not closely related) species sterrhopteron York Botanical Garden, pers. with similar nesting and foraging comm.). Although we were unable habits to Wetmorethraupis Since its discovery7 in 1963, to examine it closely, on this or (i.e., Aphelocoma, Cyanocorax, Orange-throated Tanager subsequent visits, it appeared to Cyanolyca (Corvidae), Sericossypha Wetmorethraupis sterrhopteron be an open-cup nest, supported (Thraupidae), we believe that stands as one of the most from below by the woody rachis incubation was underway on distinctive and striking new bird of a palm frond, with little or no 16–19 February. -

Reproductive Behavior of the Red-Crested Finch Coryphospingus Cucullatus (Aves: Thraupidae) in Southeastern Brazil
ZOOLOGIA 33(4): e20160071 ISSN 1984-4689 (online) www.scielo.br/zool BEHAVIOR Reproductive behavior of the Red-crested Finch Coryphospingus cucullatus (Aves: Thraupidae) in southeastern Brazil Paulo V.Q. Zima1 & Mercival R. Francisco2* 1Programa de Pós-graduação em Ecologia e Recursos Naturais, Universidade Federal de São Carlos. Rodovia Wash- ington Luís, km 235, 13565-905 São Carlos, SP, Brazil. 2Departamento de Ciências Ambientais, Universidade Federal de São Carlos, Campus Sorocaba. Rodovia João Leme dos Santos, km 110, 18052-780 Sorocaba, SP, Brazil. *Corresponding author. E-mail: [email protected] ABSTRACT. Several behavioral aspects of the Red-crested Finch Coryphospingus cucullatus (Statius Müller, 1776) are poorly studied. Here we provide reproductive information on 16 active nests. This information may be valuable to elucidate the phylogenetic relationships of this bird, and to design plans to manage it. Nesting activities occurred from October to February. Clutches consisted of two to three eggs (2.06 ± 0.25), which were laid on consecutive days. Incubation usually started the morning the females laid their last egg and lasted 11.27 ± 0.47 days. Hatching was synchronous, or happened at a one-day interval. The nestling stage lasted 12 ± 0.89 days. Only females incubated the eggs and they fed the young more often than the males did. Overall nesting success, from incubation to fledging, was 28.2%. Nest architecture and egg color proved to be diagnostic characteristics of Coryphospingus, supporting its maintenance as a distinct genus within the recently proposed sub-family Tachyphoninae. Red-crested Finches showed a preference for certain nesting sites, i.e., forest borders or a Cerrado in late regeneration stage.