Molecular Phylogenetics and Evolution 75 (2014) 41–77
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Birds of Reserva Ecológica Guapiaçu (REGUA)
Cotinga 33 The birds of Reserva Ecológica Guapiaçu (REGUA), Rio de Janeiro, Brazil Leonardo Pimentel and Fábio Olmos Received 30 September 2009; final revision accepted 15 December 2010 Cotinga 33 (2011): OL 8–24 published online 16 March 2011 É apresentada uma lista da avifauna da Reserva Ecológica de Guapiaçu (REGUA), uma reserva privada de 6.500 ha localizada no município de Cachoeiras de Macacu, vizinha ao Parque Estadual dos Três Picos, Estação Ecológica do Paraíso e Parque Nacional da Serra dos Órgãos, parte de um dos maiores conjuntos protegidos do Estado do Rio de Janeiro. Foram registradas um total de 450 espécies de aves, das quais 63 consideradas de interesse para conservação, como Leucopternis lacernulatus, Harpyhaliaetus coronatus, Triclaria malachitacea, Myrmotherula minor, Dacnis nigripes, Sporophila frontalis e S. falcirostris. A reserva também está desenvolvendo um projeto de reintrodução dos localmente extintos Crax blumembachii e Aburria jacutinga, e de reforço das populações locais de Tinamus solitarius. The Atlantic Forest of eastern Brazil and Some information has been published on neighbouring Argentina and Paraguay is among the birds of lower (90–500 m) elevations in the the most imperilled biomes in the world. At region10,13, but few areas have been subject to least 188 bird species are endemic to it, and 70 long-term surveys. Here we present the cumulative globally threatened birds occur there, most of them list of a privately protected area, Reserva Ecológica endemics4,8. The Atlantic Forest is not homogeneous Guapiaçu (REGUA), which includes both low-lying and both latitudinal and longitudinal gradients parts of the Serra dos Órgãos massif and nearby account for diverse associations of discrete habitats higher ground, now mostly incorporated within and associated bird communities. -

Cross-Temporal Analysis of Genetic Diversity in the Endangered Medium Tree Finch (Camarhynchus Pauper) and Closely Related Darwin’S Finches
Cross-Temporal Analysis of Genetic Diversity in the Endangered Medium Tree Finch (Camarhynchus pauper) and Closely Related Darwin’s Finches By Colleen Metzger B.S., Juniata College, 2009 A thesis submitted to the Graduate School of the University of Cincinnati Department of Biological Sciences In partial fulfillment of the requirements For the degree of Master of Science Committee Chair: Kenneth Petren, Ph.D. November 2012 Abstract Natural history collections can provide a direct view of past genotypes, which allows greater insight into evolutionary processes that are relevant for conservation and management. However, few studies have used broad surveys of multilocus genotypes from the past to address the wide range of processes that can affect conservation planning of a species today. Therefore, we assessed the history and status of the critically endangered medium tree finch, Camarhynchus pauper, an endemic finch of the Galápagos Islands. Using ancient DNA techniques, we quantified cross-temporal genetic change for 16 microsatellite loci in this species and its relatives. We tested the hypothesis that C. pauper has undergone a recent reduction in population size and loss of genetic diversity, and evaluated the hypothesis that C. pauper is genetically distinct from its two closest relatives, C. parvulus and C. psittacula. We assessed whether decline in C. pauper has led to increased hybridization with other species and evaluated a long-standing hypothesis of its origin from C. psittacula on another island using genetic distances, assignment tests, and migration analyses. Genetic diversity declined significantly in C. pauper over time, and several other tree finch populations showed similar losses of genetic diversity. -
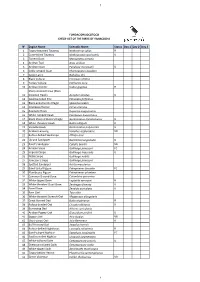
N° English Name Scientific Name Status Day 1
1 FUNDACIÓN JOCOTOCO CHECK-LIST OF THE BIRDS OF YANACOCHA N° English Name Scientific Name Status Day 1 Day 2 Day 3 1 Tawny-breasted Tinamou Nothocercus julius R 2 Curve-billed Tinamou Nothoprocta curvirostris U 3 Torrent Duck Merganetta armata 4 Andean Teal Anas andium 5 Andean Guan Penelope montagnii U 6 Sickle-winged Guan Chamaepetes goudotii 7 Cattle Egret Bubulcus ibis 8 Black Vulture Coragyps atratus 9 Turkey Vulture Cathartes aura 10 Andean Condor Vultur gryphus R Sharp-shinned Hawk (Plain- 11 breasted Hawk) Accipiter striatus U 12 Swallow-tailed Kite Elanoides forficatus 13 Black-and-chestnut Eagle Spizaetus isidori 14 Cinereous Harrier Circus cinereus 15 Roadside Hawk Rupornis magnirostris 16 White-rumped Hawk Parabuteo leucorrhous 17 Black-chested Buzzard-Eagle Geranoaetus melanoleucus U 18 White-throated Hawk Buteo albigula R 19 Variable Hawk Geranoaetus polyosoma U 20 Andean Lapwing Vanellus resplendens VR 21 Rufous-bellied Seedsnipe Attagis gayi 22 Upland Sandpiper Bartramia longicauda R 23 Baird's Sandpiper Calidris bairdii VR 24 Andean Snipe Gallinago jamesoni FC 25 Imperial Snipe Gallinago imperialis U 26 Noble Snipe Gallinago nobilis 27 Jameson's Snipe Gallinago jamesoni 28 Spotted Sandpiper Actitis macularius 29 Band-tailed Pigeon Patagoienas fasciata FC 30 Plumbeous Pigeon Patagioenas plumbea 31 Common Ground-Dove Columbina passerina 32 White-tipped Dove Leptotila verreauxi R 33 White-throated Quail-Dove Zentrygon frenata U 34 Eared Dove Zenaida auriculata U 35 Barn Owl Tyto alba 36 White-throated Screech-Owl Megascops -

Bulletin Biological Assessment Boletín RAP Evaluación Biológica
Rapid Assessment Program Programa de Evaluación Rápida Evaluación Biológica Rápida de Chawi Grande, Comunidad Huaylipaya, Zongo, La Paz, Bolivia RAP Bulletin A Rapid Biological Assessment of of Biological Chawi Grande, Comunidad Huaylipaya, Assessment Zongo, La Paz, Bolivia Boletín RAP de Evaluación Editores/Editors Biológica Claudia F. Cortez F., Trond H. Larsen, Eduardo Forno y Juan Carlos Ledezma 70 Conservación Internacional Museo Nacional de Historia Natural Gobierno Autónomo Municipal de La Paz Rapid Assessment Program Programa de Evaluación Rápida Evaluación Biológica Rápida de Chawi Grande, Comunidad Huaylipaya, Zongo, La Paz, Bolivia RAP Bulletin A Rapid Biological Assessment of of Biological Chawi Grande, Comunidad Huaylipaya, Assessment Zongo, La Paz, Bolivia Boletín RAP de Evaluación Editores/Editors Biológica Claudia F. Cortez F., Trond H. Larsen, Eduardo Forno y Juan Carlos Ledezma 70 Conservación Internacional Museo Nacional de Historia Natural Gobierno Autónomo Municipal de La Paz The RAP Bulletin of Biological Assessment is published by: Conservation International 2011 Crystal Drive, Suite 500 Arlington, VA USA 22202 Tel: +1 703-341-2400 www.conservation.org Cover Photos: Trond H. Larsen (Chironius scurrulus). Editors: Claudia F. Cortez F., Trond H. Larsen, Eduardo Forno y Juan Carlos Ledezma Design: Jaime Fernando Mercado Murillo Map: Juan Carlos Ledezma y Veronica Castillo ISBN 978-1-948495-00-4 ©2018 Conservation International All rights reserved. Conservation International is a private, non-proft organization exempt from federal income tax under section 501c(3) of the Internal Revenue Code. The designations of geographical entities in this publication, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of Conservation International or its supporting organizations concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. -

Diversity and Natural History of Birds in Green Urban Areas of the City of Quito, Ecuador (America)
Diversity and natural history of birds in green urban areas of the city of Quito, Ecuador (America) Diego F. Cisneros-Heredia1,2,3* & Eliana Montenegro1 1 Universidad San Francisco de Quito, Colegio de Ciencias Biológicas y Ambientales, Laboratorio de Zoología Terrestre. Casilla Postal 17-1200-841, Quito, Ecuador 2 King’s College London, Department of Geography, Strand, London WC2R 2LS, UK 3 Aves&Conservación / BirdLife Ecuador, Quito, Ecuador * E-mail: [email protected] Abstract The diversity and ecology of urban bird communities have been extensively studied in Neartic and Paleartic areas, however, little is known about urban Neotropical areas. Quito, capital city of Ecuador, is located on a highland valley in the megadiverse tropical Andes. Founded in 1534, Quito did not increase significantly its urban area until the late 19th century, growing at an accelerated and unplanned rate during the 20th century. More than 100 species were known to inhabit in Quito at the end of the 19th century. Currently, most authors estimated that no more than 40 species occur in Quito, although no systematic bird studies have been conducted. Our research is a first approach to the avifauna of Quito, surveying the diversity living in green urban areas within the city borders. We used two field methodologies, i.e. line transects and point counts, to survey 16 green urban areas over 12 months. We recorded 65 species of birds, belonging to 20 families and 9 orders. Three species were the most common and frequent: Eared Dove Zenaida auriculata, Rufous-collared Sparrow Zonotrichia capensis, and Great Thrush Turdus fuscater, being omnivores and granivores adapted to anthropic habitats with low ecological complexity. -

REGUA Bird List July 2020.Xlsx
Birds of REGUA/Aves da REGUA Updated July 2020. The taxonomy and nomenclature follows the Comitê Brasileiro de Registros Ornitológicos (CBRO), Annotated checklist of the birds of Brazil by the Brazilian Ornithological Records Committee, updated June 2015 - based on the checklist of the South American Classification Committee (SACC). Atualizado julho de 2020. A taxonomia e nomenclatura seguem o Comitê Brasileiro de Registros Ornitológicos (CBRO), Lista anotada das aves do Brasil pelo Comitê Brasileiro de Registros Ornitológicos, atualizada em junho de 2015 - fundamentada na lista do Comitê de Classificação da América do Sul (SACC). -

Ornithological Surveys in Serranía De Los Churumbelos, Southern Colombia
Ornithological surveys in Serranía de los Churumbelos, southern Colombia Paul G. W . Salaman, Thomas M. Donegan and Andrés M. Cuervo Cotinga 12 (1999): 29– 39 En el marco de dos expediciones biológicos y Anglo-Colombian conservation expeditions — ‘Co conservacionistas anglo-colombianas multi-taxa, s lombia ‘98’ and the ‘Colombian EBA Project’. Seven llevaron a cabo relevamientos de aves en lo Serranía study sites were investigated using non-systematic de los Churumbelos, Cauca, en julio-agosto 1988, y observations and standardised mist-netting tech julio 1999. Se estudiaron siete sitios enter en 350 y niques by the three authors, with Dan Davison and 2500 m, con 421 especes registrados. Presentamos Liliana Dávalos in 1998. Each study site was situ un resumen de los especes raros para cada sitio, ated along an altitudinal transect at c. 300- incluyendo los nuevos registros de distribución más m elevational steps, from 350–2500 m on the Ama significativos. Los resultados estabilicen firme lo zonian slope of the Serranía. Our principal aim was prioridad conservacionista de lo Serranía de los to allow comparisons to be made between sites and Churumbelos, y aluco nos encontramos trabajando with other biological groups (mammals, herptiles, junto a los autoridades ambientales locales con insects and plants), and, incorporating geographi cuiras a lo protección del marcizo. cal and anthropological information, to produce a conservation assessment of the region (full results M e th o d s in Salaman et al.4). A sizeable part of eastern During 14 July–17 August 1998 and 3–22 July 1999, Cauca — the Bota Caucana — including the 80-km- ornithological surveys were undertaken in Serranía long Serranía de los Churumbelos had never been de los Churumbelos, Department of Cauca, by two subject to faunal surveys. -

Ficha Informativa De Los Humedales De Ramsar (FIR) Versión 2009-2012
Ficha Informativa de los Humedales de Ramsar (FIR) versión 2009-2012 1. Nombre y dirección del compilador de la Ficha: PARA USO INTERNO DE LA OFICINA DE RAMSAR . DD MM YY Sandro Menezes Silva Conservação Internacional (CI-Brasil) R. Paraná, 32 CEP-79020-290 Designation date Site Reference Number Campo Grande - MS – Brasil [email protected] Tel: +55(67) 3326-0002 Fax: +55(67) 3326-8737 2. Fecha en que la Ficha se llenó /actualizó : Julio 2008 3. País: Brasil 4. Nombre del sitio Ramsar: Reserva Particular del Patrimonio Natural (RPPN) “Fazenda Rio Negro” 5. Designación de nuevos sitios Ramsar o actualización de los ya existentes: Esta FIR es para (marque una sola casilla) : a) Designar un nuevo sitio Ramsar o b) Actualizar información sobre un sitio Ramsar existente 6. Sólo para las actualizaciones de FIR, cambios en el sitio desde su designación o anterior actualización: 7. Mapa del sitio: a) Se incluye un mapa del sitio, con límites claramente delineados, con el siguiente formato: i) versión impresa (necesaria para inscribir el sitio en la Lista de Ramsar): Anexo 1 ; ii ) formato electrónico (por ejemplo, imagen JPEG o ArcView) iii) un archivo SIG con tablas de atributos y vectores georreferenciados sobre los límites del sitio b) Describa sucintamente el tipo de delineación de límites aplicado: El límite del Sitio Ramsar es el mismo de la RPPN Fazenda Rio Negro, reconocida oficialmente como área protegida por el gobierno de la Provincia de Mato Grosso 8. Coordenadas geográficas (latitud / longitud, en grados y minutos): Lat 19°33'2.78"S / long 56°13'27.93"O (coordenadas de la sede de la hacienda) 9. -

Dacninae Species Tree, Part I
Dacninae I: Nemosiini, Conirostrini, & Diglossini Hooded Tanager, Nemosia pileata Cherry-throated Tanager, Nemosia rourei Nemosiini Blue-backed Tanager, Cyanicterus cyanicterus White-capped Tanager, Sericossypha albocristata Scarlet-throated Tanager, Sericossypha loricata Bicolored Conebill, Conirostrum bicolor Pearly-breasted Conebill, Conirostrum margaritae Chestnut-vented Conebill, Conirostrum speciosum Conirostrini White-eared Conebill, Conirostrum leucogenys Capped Conebill, Conirostrum albifrons Giant Conebill, Conirostrum binghami Blue-backed Conebill, Conirostrum sitticolor White-browed Conebill, Conirostrum ferrugineiventre Tamarugo Conebill, Conirostrum tamarugense Rufous-browed Conebill, Conirostrum rufum Cinereous Conebill, Conirostrum cinereum Stripe-tailed Yellow-Finch, Pseudochloris citrina Gray-hooded Sierra Finch, Phrygilus gayi Patagonian Sierra Finch, Phrygilus patagonicus Peruvian Sierra Finch, Phrygilus punensis Black-hooded Sierra Finch, Phrygilus atriceps Gough Finch, Rowettia goughensis White-bridled Finch, Melanodera melanodera Yellow-bridled Finch, Melanodera xanthogramma Inaccessible Island Finch, Nesospiza acunhae Nightingale Island Finch, Nesospiza questi Wilkins’s Finch, Nesospiza wilkinsi Saffron Finch, Sicalis flaveola Grassland Yellow-Finch, Sicalis luteola Orange-fronted Yellow-Finch, Sicalis columbiana Sulphur-throated Finch, Sicalis taczanowskii Bright-rumped Yellow-Finch, Sicalis uropigyalis Citron-headed Yellow-Finch, Sicalis luteocephala Patagonian Yellow-Finch, Sicalis lebruni Greenish Yellow-Finch, -

Costa Rica: the Introtour | July 2017
Tropical Birding Trip Report Costa Rica: The Introtour | July 2017 A Tropical Birding SET DEPARTURE tour Costa Rica: The Introtour July 15 – 25, 2017 Tour Leader: Scott Olmstead INTRODUCTION This year’s July departure of the Costa Rica Introtour had great luck with many of the most spectacular, emblematic birds of Central America like Resplendent Quetzal (photo right), Three-wattled Bellbird, Great Green and Scarlet Macaws, and Keel-billed Toucan, as well as some excellent rarities like Black Hawk- Eagle, Ochraceous Pewee and Azure-hooded Jay. We enjoyed great weather for birding, with almost no morning rain throughout the trip, and just a few delightful afternoon and evening showers. Comfortable accommodations, iconic landscapes, abundant, delicious meals, and our charismatic driver Luís enhanced our time in the field. Our group, made up of a mix of first- timers to the tropics and more seasoned tropical birders, got along wonderfully, with some spying their first-ever toucans, motmots, puffbirds, etc. on this trip, and others ticking off regional endemics and hard-to-get species. We were fortunate to have several high-quality mammal sightings, including three monkey species, Derby’s Wooly Opossum, Northern Tamandua, and Tayra. Then there were many www.tropicalbirding.com +1-409-515-9110 [email protected] Page Tropical Birding Trip Report Costa Rica: The Introtour | July 2017 superb reptiles and amphibians, among them Emerald Basilisk, Helmeted Iguana, Green-and- black and Strawberry Poison Frogs, and Red-eyed Leaf Frog. And on a daily basis we saw many other fantastic and odd tropical treasures like glorious Blue Morpho butterflies, enormous tree ferns, and giant stick insects! TOP FIVE BIRDS OF THE TOUR (as voted by the group) 1. -

UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS Departamento De Zoología Y Antropología Física
UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS Departamento de Zoología y Antropología Física TESIS DOCTORAL Diversidad y especificidad de simbiontes en aves neotropicales Diversity and host specificity of symbionts in neotropical birds MEMORIA PARA OPTAR AL GRADO DE DOCTOR PRESENTADA POR Michaël André Jean Moens Directores Javier Pérez Tris Laura Benítez Rico Madrid, 2017 © Michaël André Jean Moens, 2016 Diversidad y Especificidad de Simbiontes en Aves Neotropicales (Diversity and Host Specificity of Symbionts in Neotropical birds.) Tesis doctoral de: Michaël André Jean Moens Directores : Javier Pérez Tris Laura Benítez Rico Madrid, 2016 © Michaël André Jean Moens, 2016 Cover Front Chestnut-breasted Coronet (Boissoneaua matthewsii) Taken at the San Isidro Reserve, Ecuador. Copyright © Jaime Culebras Cover Back Royal Flycatcher (Onychorhynchus coronatus) Taken at the Nouragues Reserve, French Guiana. Copyright © Borja Milá UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS DEPARTAMENTO DE ZOOLOGÍA Y ANTROPOLOGÍA FÍSICA Diversidad y Especificidad de Simbiontes en Aves Neotropicales (Diversity and Host Specificity of Symbionts in Neotropical birds.) Tesis doctoral de: Michaël André Jean Moens Directores: Javier Pérez Tris Laura Benítez Rico Madrid, 2016 © Michaël André Jean Moens, 2016 UNIVERSIDAD COMPLUTENSE DE MADRID FACULTAD DE CIENCIAS BIOLÓGICAS DEPARTAMENTO DE ZOOLOGÍA Y ANTROPOLOGÍA FÍSICA Diversidad y Especificidad de Simbiontes en Aves Neotropicales (Diversity and Host Specificity of Symbionts -

Southeastern Brazil: Best of the Atlantic Forest
SOUTHEASTERN BRAZIL: BEST OF THE ATLANTIC FOREST OCTOBER 8–22, 2017 A trip first, the rarely seen Buff-fronted Owl – Photo: Andrew Whittaker LEADER : ANDREW WHITTAKER LIST COMPILED BY : ANDREW WHITTAKER VICTOR EMANUEL NATURE TOURS , INC . 2525 WALLINGWOOD DRIVE , SUITE 1003 AUSTIN , TEXAS 78746 WWW .VENTBIRD .COM SOUTHEASTERN BRAZIL: BEST OF THE ATLANTIC FOREST OCTOBER 8–22, 2017 By Andrew Whittaker Once again, our Brazilian flagship tour visiting the lovely southeast rocked, delivering a bonanza of Atlantic Forest endemics, spectacular scenery, all-around great birding, and wonderful Brazilian cuisine that we have come to expect from this fantastic biologically rich region. First and foremost we tallied 391 species , a whopping 140 of which were regional and/or Brazilian endemics! These figures become all the more impressive when you consider that many of the wider ranging species not included as “endemics” in the preceding tallies are represented in southeast Brazil by distinctive subspecies endemic to the Atlantic Forest region, and that at least 15–25 of these subspecies that we recorded during our tours are likely to be elevated to separate species status in the near future! Beginning in São Paulo, our first destination was Intervales State Park, my own personal favorite among the many great birding spots included in our southeast Brazil trip. Intervales never fails to deliver a huge serving of Atlantic Forest endemics and just plain fantastic birding experiences, and such was the case again this trip. We began the first evening with one of my personal highlights, the fabulous and incredibly cooperative male Long-trained Nightjar . In fact, the male Long-trained Nightjars put on a show for us on two consecutive nights, treating us to multiple close passes, with two males chasing each other just above our heads.