Exploring Deletion Strategies for the Bond-Tree in Multidimensional Non-Ordered Discrete Data Spaces
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Suffix Trees for Fast Sensor Data Forwarding
Suffix Trees for Fast Sensor Data Forwarding Jui-Chieh Wub Hsueh-I Lubc Polly Huangac Department of Electrical Engineeringa Department of Computer Science and Information Engineeringb Graduate Institute of Networking and Multimediac National Taiwan University [email protected], [email protected], [email protected] Abstract— In data-centric wireless sensor networks, data are alleviates the effort of node addressing and address reconfig- no longer sent by the sink node’s address. Instead, the sink uration in large-scale mobile sensor networks. node sends an explicit interest for a particular type of data. The source and the intermediate nodes then forward the data Forwarding in data-centric sensor networks is particularly according to the routing states set by the corresponding interest. challenging. It involves matching of the data content, i.e., This data-centric style of communication is promising in that it string-based attributes and values, instead of numeric ad- alleviates the effort of node addressing and address reconfigura- dresses. This content-based forwarding problem is well studied tion. However, when the number of interests from different sinks in the domain of publish-subscribe systems. It is estimated increases, the size of the interest table grows. It could take 10s to 100s milliseconds to match an incoming data to a particular in [3] that the time it takes to match an incoming data to a interest, which is orders of mangnitude higher than the typical particular interest ranges from 10s to 100s milliseconds. This transmission and propagation delay in a wireless sensor network. processing delay is several orders of magnitudes higher than The interest table lookup process is the bottleneck of packet the propagation and transmission delay. -

Design and Analysis of Data Structures
Contents 1 Introduction and Review 1 1.1 Welcome! . .1 1.2 Tick Tock, Tick Tock . .2 1.3 Classes of Computational Complexity . .6 1.4 The Fuss of C++ . 12 1.5 Random Numbers . 30 1.6 Bit-by-Bit . 33 1.7 The Terminal-ator . 38 1.8 Git, the \Undo" Button of Software Development . 43 2 Introductory Data Structures 47 2.1 Array Lists . 47 2.2 Linked Lists . 53 2.3 Skip Lists . 59 2.4 Circular Arrays . 67 2.5 Abstract Data Types . 75 2.6 Deques . 76 2.7 Queues . 79 2.8 Stacks . 81 2.9 And the Iterators Gonna Iterate-ate-ate . 84 3 Tree Structures 91 3.1 Lost in a Forest of Trees . 91 3.2 Heaps . 99 3.3 Binary Search Trees . 105 3.4 BST Average-Case Time Complexity . 112 3.5 Randomized Search Trees . 117 3.6 AVL Trees . 125 3.7 Red-Black Trees . 136 3.8 B-Trees . 145 3.9 B+ Trees . 156 i ii CONTENTS 4 Introduction to Graphs 167 4.1 Introduction to Graphs . 167 4.2 Graph Representations . 173 4.3 Algorithms on Graphs: Breadth First Search . 177 4.4 Algorithms on Graphs: Depth First Search . 181 4.5 Dijkstra's Algorithm . 185 4.6 Minimum Spanning Trees . 190 4.7 Disjoint Sets . 197 5 Hashing 209 5.1 The Unquenched Need for Speed . 209 5.2 Hash Functions . 210 5.3 Introduction to Hash Tables . 215 5.4 Probability of Collisions . 221 5.5 Collision Resolution: Open Addressing . 227 5.6 Collision Resolution: Closed Addressing . -

Final Review
Final Review 1 Final Exam (Out of 70) • Part A: Basic knowledge of data structures – 20 points – MulEple choice • Part B: Applicaon, Comparison and Implementaon of the data structures – 20 points – Apply supported operaons (like find and insert) to data structures we have covered like: BST, AVL, RBTs, MulEway trie, Ternary Trie, B trees, skip lists. Also essenEal concepts in Huffman codes • Part C: Simulang algorithms and run Eme analysis – 15 points – Graph algorithms: BFS, DFS, Dijkstra, Prims’, Kruskals’. Also union find • Part D: C++ and programming assignments – 15 points – Short answer 2 Final Exam Practice Questions CSE 100 (Fall 2014) CSE Department University of California, San Diego Part A: The Basics This section tests your basic knowledge of data structures via multiple choice questions. Sample questions include all the iclicker and reading quiz questions covered in class. Please make sure you review them. Part B: Application, Comparison and Implementation This section tests how well you understand what goes on under the hood of the data structures covered during the course, their strengths and weaknesses and their applications. The format is short answers and fill in the blanks. 1. B-trees. (a) Construct a 2-3 tree by inserting the following keys in the order shown: 10, 15, 20, 25, 17, 30. You can check your answers and experiment with trees of your own design at the following web sites: https://www.cs.usfca.edu/˜galles/visualization/BTree.html B-trees http://ats.oka.nu/b-tree/b-tree.manual.html (b) Which of the following are legal 2,3 trees (B tree of order 3)? For a tree that is not a valid 2,3 tree, state a reason why. -

Practical Concurrent Traversals in Search Trees
Practical Concurrent Traversals in Search Trees Dana Drachsler-Cohen∗ Martin Vechev Eran Yahav ETH Zurich, Switzerland ETH Zurich, Switzerland Technion, Israel [email protected] [email protected] [email protected] Abstract efficiently remains a difficult11 task[ ]. This task is partic- Operations of concurrent objects often employ optimistic ularly challenging for search trees whose traversals may concurrency-control schemes that consist of a traversal fol- be performed concurrently with modifications that relocate lowed by a validation step. The validation checks if concur- nodes. Thus, a major challenge in designing a concurrent rent mutations interfered with the traversal to determine traversal operation is to ensure that target nodes are not if the operation should proceed or restart. A fundamental missed, even if they are relocated. This challenge is ampli- challenge is to discover a necessary and sufficient validation fied when a concurrent data structure is optimized forread check that has to be performed to guarantee correctness. operations and traversals are expected to complete without In this paper, we show a necessary and sufficient condi- costly synchronization primitives (e.g., locks, CAS). Recent tion for validating traversals in search trees. The condition work offers various concurrent data structures optimized for relies on a new concept of succinct path snapshots, which are lightweight traversals [1, 2, 4, 6–10, 12–14, 18, 19, 22, 25, 27]. derived from and embedded in the structure of the tree. We However, they are mostly specialized solutions, supporting leverage the condition to design a general lock-free mem- standard operations, that cannot be applied directly to other bership test suitable for any search tree. -

Multivariate Normal Limit Laws for the Numbers of Fringe Subtrees in M-Ary Search Trees and Preferential Attachment Trees
Multivariate normal limit laws for the numbers of fringe subtrees in m-ary search trees and preferential attachment trees Cecilia Holmgren∗ Svante Jansony Matas Sileikisˇ Department of Mathematics Department of Applied Mathematics Uppsala University Faculty of Mathematics and Physics PO Box 480 Charles University 751 06 Uppsala, Sweden Malostransk´en´am.25 fcecilia.holmgren,[email protected] 118 00 Praha, Czech Republic [email protected] Submitted: Aug 12, 2016; Accepted: Jun 22, 2017; Published: Jun 30, 2017 Mathematics Subject Classifications: 60C05; 05C05; 05C80; 60F05; 68P05; 68P10 Abstract We study fringe subtrees of random m-ary search trees and of preferential attach- ment trees, by putting them in the context of generalised P´olya urns. In particular we show that for the random m-ary search trees with m 6 26 and for the linear preferential attachment trees, the number of fringe subtrees that are isomorphic to an arbitrary fixed tree T converges to a normal distribution; more generally, we also prove multivariate normal distribution results for random vectors of such numbers for different fringe subtrees. Furthermore, we show that the number of protected nodes in random m-ary search trees for m 6 26 has asymptotically a normal distribution. Keywords: Random trees; Fringe trees; Normal limit laws; P´olya urns; m-ary search trees; Preferential attachment trees; Protected nodes 1 Introduction The main focus of this paper is to consider fringe subtrees of random m-ary search trees and of general preferential attachment trees (including the random recursive tree); these random trees are defined in Section2. Recall that a fringe subtree is a subtree consisting of some node and all its descendants, see Aldous [1] for a general theory, and note that ∗Partly supported by the Swedish Research Council. -

Package 'Tstr'
Package ‘TSTr’ October 31, 2015 Type Package Title Ternary Search Tree for Auto-Completion and Spell Checking Version 1.2 Date 2015-10-07 Author Ricardo Merino [aut, cre], Samantha Fernandez [ctb] Maintainer Ricardo Merino <[email protected]> Description A ternary search tree is a type of prefix tree with up to three children and the abil- ity for incremental string search. The package uses this ability for word auto- completion and spell checking. Includes a dataset with the 10001 most frequent English words. License GPL-2 LazyData yes Depends R (>= 3.2.0) Imports stringr, stringdist, stats, data.table Suggests knitr VignetteBuilder knitr NeedsCompilation no R topics documented: TSTr-package . .2 addToTree . .2 addWord . .3 completeWord . .4 dimTree . .4 newTree . .5 PNcheck . .6 SDcheck . .7 SDkeeper . .8 searchWord . .9 XMIwords . .9 Index 10 1 2 addToTree TSTr-package Ternary Search Tree for Auto-Completion and Spell Checking Description A ternary search tree is a type of prefix tree with up to three children and the ability for incremental string search. The package uses this ability for word auto-completion and spell checking. Includes a dataset with the 10001 most frequent English words. Details This package can be used to create a ternary search tree, more space efficient compared to stan- dard prefix trees. Common applications for ternary search trees include spell-checking and auto- completion. Author(s) Ricardo Merino [aut, cre], Samantha Fernandez [ctb] Maintainer: Ricardo Merino <[email protected]> References https://en.wikipedia.org/wiki/Ternary_search_tree See Also newTree addToTree Adds a set of strings to a ternary search tree Description Updates a ternary search tree adding the words or strings from the input Usage addToTree(tree, input) Arguments tree an existing ternary search tree to add words to. -

Implementasi Algoritma Ternary Search Tree Dan Teknologi Grafis Berbasis Vektor Untuk Interpretasi Alfabet Pitman Shorthand
Implementasi Algoritma Ternary Search Tree dan Teknologi Grafis Berbasis Vektor untuk Interpretasi Alfabet Pitman Shorthand 1) Irwan Sembiring, 2) Theophilus Wellem, 3) Gloria Saripah Patara Fakultas Teknologi Informasi Universitas Kristen Satya Wacana Jl. Diponegoro 52 – 60, Salatiga 50711, Indonesia Email : 1) [email protected], 2) [email protected], 3) [email protected] Abstract Pitman shorthand system is one of the most popular of nowadays shorthand systems. It has been taught and widely used throughout the world to help the stenographers in improving the writing speed. In this research, a Pitman shorthand’s alphabets interpretation mobile system is established purposely to ease the stenographers in interpreting the shorthand script into English text. The outlines are interpreted using dictionary-based approach. This system can display alphabet list, so that it also can be used by the students who wish to learn this shorthand system. All symbols are rendered with JSR-226 (Scalable Vector Graphics API). The system implements Ternary Search Tree (TST) algorithm to improve data searching speed. According to the experiments, the system can successfully interpret inputted alphabets up to 71.95%. Key Words: Pitman Shorthand, Stenographer, Dictionary-based , Approach, JSR-226 (Scalable Vector Graphics API), Ternary Search Tree. 1. Pendahuluan Pitman Shorthand merupakan sistem penulisan cepat yang dikembangkan oleh Sir Isaac Pitman (1813-1897). Sistem penulisan cepat ini dipublikasikan pada tahun 1837. Pitman Shorthand adalah sistem fonetis yang menggunakan simbol-simbol untuk berbagai bunyi dalam bahasa [1]. Pemakaian simbol untuk merepresentasikan bunyi tertentu dimaksudkan untuk mempercepat penulisan suatu kata. Hal ini disebabkan suatu simbol dapat menggantikan pemakaian beberapa alfabet Latin yang sering digunakan dalam suatu kata. -

Burst Tries: a Fast, Efficient Data Structure for String Keys
Burst Tries: A Fast, Efficient Data Structure for String Keys Ste↵en Heinz Justin Zobel⇤ Hugh E. Williams School of Computer Science and Information Technology, RMIT University GPO Box 2476V, Melbourne 3001, Australia sheinz,jz,hugh @cs.rmit.edu.au { } Abstract Many applications depend on efficient management of large sets of distinct strings in memory. For example, during index construction for text databases a record is held for each distinct word in the text, containing the word itself and information such as counters. We propose a new data structure, the burst trie, that has significant advantages over existing options for such applications: it requires no more memory than a binary tree; it is as fast as a trie; and, while not as fast as a hash table, a burst trie maintains the strings in sorted or near-sorted order. In this paper we describe burst tries and explore the parameters that govern their performance. We experimentally determine good choices of parameters, and compare burst tries to other structures used for the same task, with a variety of data sets. These experiments show that the burst trie is particularly e↵ective for the skewed frequency distributions common in text collections, and dramatically outperforms all other data structures for the task of managing strings while maintaining sort order. Keywords Tries, binary trees, splay trees, string data structures, text databases. 1 Introduction Many computing applications involve management of large sets of distinct strings. For example, vast quantities of information are held as text documents, ranging from news archives, law collections, and business reports to repositories of documents gathered from the web. -

Randomized Ternary Search Tries
Randomized Ternary Search Tries Nicolai Diethelm Abstract A simple method for maintaining balance in ternary search tries is presented. The new kind of self- balancing ternary search trie, called an r-trie, generalizes the balancing strategy of Aragon and Seidel’s randomized binary search trees (treaps). This means that the shape of an r-trie for a string set S is a random variable with the same probability distribution as a ternary search trie built by inserting the strings of S in random order. It is shown that searching, inserting, or deleting a string of length k in an r-trie for n strings takes at most O(k + log n) time with high probability, no matter from which sequence of insertions and deletions of strings the r-trie results. 1 Introduction In computer science, tries are an important data structure for storing character strings. If viewed as an abstract structure, the trie for a set S of strings can be defined to be the smallest ordered tree such that each node except the root is labeled with a character and each string in S is spelled out by the characters on a path from the root to a node. Each node in the trie corresponds to a prefix of the strings in S (namely the prefix spelled out by the path from the root to the node) and vice versa. A node that corresponds to a string in S is called a terminal node. Any data associated with a string in S is stored at the terminal node of the string. -

INDIAN INSTITUTE of TECHNOLOGY KHARAGPUR Stamp / Signature of the Invigilator
INDIAN INSTITUTE OF TECHNOLOGY KHARAGPUR Stamp / Signature of the Invigilator EXAMINATION ( End Semester ) SEMESTER ( Spring ) Roll Number Section Name Subject Number C S 2 1 0 0 3 Subject Name Algorithms – I Department / Center of the Student Additional sheets Important Instructions and Guidelines for Students 1. You must occupy your seat as per the Examination Schedule/Sitting Plan. 2. Do not keep mobile phones or any similar electronic gadgets with you even in the switched off mode. 3. Loose papers, class notes, books or any such materials must not be in your possession, even if they are irrelevant to the subject you are taking examination. 4. Data book, codes, graph papers, relevant standard tables/charts or any other materials are allowed only when instructed by the paper-setter. 5. Use of instrument box, pencil box and non-programmable calculator is allowed during the examination. However, exchange of these items or any other papers (including question papers) is not permitted. 6. Write on both sides of the answer script and do not tear off any page. Use last page(s) of the answer script for rough work. Report to the invigilator if the answer script has torn or distorted page(s). 7. It is your responsibility to ensure that you have signed the Attendance Sheet. Keep your Admit Card/Identity Card on the desk for checking by the invigilator. 8. You may leave the examination hall for wash room or for drinking water for a very short period. Record your absence from the Examination Hall in the register provided. Smoking and the consumption of any kind of beverages are strictly prohibited inside the Examination Hall. -
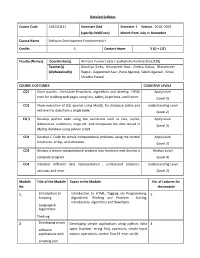
Detailed Syllabus Course Code 15B11CI111 Semester
Detailed Syllabus Course Code 15B11CI111 Semester Odd Semester I. Session 2018 -2019 (specify Odd/Even) Month from July to December Course Name Software Development Fundamentals-I Credits 4 Contact Hours 3 (L) + 1(T) Faculty (Names) Coordinator(s) Archana Purwar ( J62) + Sudhanshu Kulshrestha (J128) Teacher(s) Adwitiya Sinha, Amanpreet Kaur, Chetna Dabas, Dharamveer (Alphabetically) Rajput, Gaganmeet Kaur, Parul Agarwal, Sakshi Agarwal , Sonal, Shradha Porwal COURSE OUTCOMES COGNITIVE LEVELS CO1 Solve puzzles , formulate flowcharts, algorithms and develop HTML Apply Level code for building web pages using lists, tables, hyperlinks, and frames (Level 3) CO2 Show execution of SQL queries using MySQL for database tables and Understanding Level retrieve the data from a single table. (Level 2) CO 3 Develop python code using the constructs such as lists, tuples, Apply Level dictionaries, conditions, loops etc. and manipulate the data stored in (Level 3) MySQL database using python script. CO4 Develop C Code for simple computational problems using the control Apply Level structures, arrays, and structure. (Level 3) CO5 Analyze a simple computational problem into functions and develop a Analyze Level complete program. (Level 4) CO6 Interpret different data representation , understand precision, Understanding Level accuracy and error (Level 2) Module Title of the Module Topics in the Module No. of Lectures for No. the module 1. Introduction to Introduction to HTML, Tagging v/s Programming, 5 Scripting Algorithmic Thinking and Problem Solving, Introductory algorithms and flowcharts Language & Algorithmic Thinking 2. Developing simple Developing simple applications using python; data 4 software types (number, string, list), operators, simple input applications with output, operations, control flow (if -else, while) scripting and visual languages 3. -
Transform Based and Search Aware Text Compression Schemes and Compressed Domain Text Retrieval
University of Central Florida STARS Electronic Theses and Dissertations, 2004-2019 2005 Transform Based And Search Aware Text Compression Schemes And Compressed Domain Text Retrieval Nan Zhang University of Central Florida Part of the Computer Sciences Commons, and the Engineering Commons Find similar works at: https://stars.library.ucf.edu/etd University of Central Florida Libraries http://library.ucf.edu This Doctoral Dissertation (Open Access) is brought to you for free and open access by STARS. It has been accepted for inclusion in Electronic Theses and Dissertations, 2004-2019 by an authorized administrator of STARS. For more information, please contact [email protected]. STARS Citation Zhang, Nan, "Transform Based And Search Aware Text Compression Schemes And Compressed Domain Text Retrieval" (2005). Electronic Theses and Dissertations, 2004-2019. 419. https://stars.library.ucf.edu/etd/419 Transform Based and Search Aware Text Compression Schemes and Compressed Domain Text Retrieval by Nan Zhang B.S. Beijing Colloge of Economics, 1990 M.S. National University of Singapore, 1998 A dissertation submitted in partial ful¯llment of the requirements for the degree of Doctor of Philosophy in the School of Computer Science in the College of College of Engineering and Computer Science at the University of Central Florida Orlando, Florida Spring Term 2005 Major Professor: Amar Mukherjee °c 2005 by Nan Zhang Abstract In recent times, we have witnessed an unprecedented growth of textual information via the Internet, digital libraries and archival text in many applications. While a good fraction of this information is of transient interest, useful information of archival value will continue to accumulate.