Upregulation of Programmed Death-1 Ligand 2 (PDCD1LG2)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Immuno-Oncology Panel 1
Immuno-Oncology panel 1 Gene Symbol Target protein name UniProt ID (& link) Modification* (56 analytes) ADA17 ADAM17 metalloprotease domain 17 P78536 *blanks mean the assay detects the ANXA1 Annexin A1 P04083 non-modified peptide sequence ANXA1 Annexin A1 P04083 ARG2 arginase, type II P78540 ATM Serine-protein kinase ATM, Ataxia telangiectasia mutated Q13315 pS2996 ATM Serine-protein kinase ATM, Ataxia telangiectasia mutated Q13315 ATM Serine-protein kinase ATM, Ataxia telangiectasia mutated Q13315 pS367 ATM Serine-protein kinase ATM, Ataxia telangiectasia mutated Q13315 C10orf54 / VISTA chromosome 10 open reading frame 54 Q9H7M9 CCL5 C-C motif chemokine ligand 5 P13501 CD14 CD14 molecule P08571 CD163 CD163 molecule Q86VB7 CD274 / PDL1 Programmed cell death 1 ligand 1 CD274 Q9NZQ7 CD33 CD33 molecule P20138 CD40/TNR5 tumor necrosis factor receptor superfamily member 5 P25942 CD40/TNR5 tumor necrosis factor receptor superfamily member 5 P25942 CD47 CD47 molecule Q08722 CD70 CD70 antigen P32970 CD74/HG2A CD74 molecule, major histocompatibility complex, class II invariant chain Q8SNA0 CEACAM8 carcinoembryonic antigen-related cell adhesion molecule 8 P31997 CX3CL1 C-X3-C motif chemokine ligand 1 P78423 CXCL10 C-X-C motif chemokine ligand 10 P02778 CXCL13 chemokine (C-X-C motif) ligand 13 O43927 ENTPD1 ectonucleoside triphosphate diphosphohydrolase 1 Q86VV3 FAS/TNR6 Fas (TNF receptor superfamily, member 6) P25445 pY291 FAS/TNR6 Fas (TNF receptor superfamily, member 6) P25445 GAPDH Glyceraldehyde-3-phosphate dehydrogenase P04406 HAVCR2 hepatitis -

Immune-Checkpoint Blockade Therapy in Lymphoma
International Journal of Molecular Sciences Review Immune-Checkpoint Blockade Therapy in Lymphoma Ayumi Kuzume 1,2, SungGi Chi 1 , Nobuhiko Yamauchi 1 and Yosuke Minami 1,* 1 Department of Hematology, National Cancer Center Hospital East, Kashiwa 277–8577, Japan; [email protected] (A.K.); [email protected] (S.C.); [email protected] (N.Y.) 2 Department of Hematology, Kameda Medical Center, Kamogawa 296–8602, Japan * Correspondence: [email protected]; Tel.: +81-4-7133-1111; Fax: +81-7133-6502 Received: 11 June 2020; Accepted: 28 July 2020; Published: 30 July 2020 Abstract: Tumor cells use immune-checkpoint pathways to evade the host immune system and suppress immune cell function. These cells express programmed cell-death protein 1 ligand 1 (PD-L1)/PD-L2, which bind to the programmed cell-death protein 1 (PD-1) present on cytotoxic T cells, trigger inhibitory signaling, and reduce cytotoxicity and T-cell exhaustion. Immune-checkpoint blockade can inhibit this signal and may serve as an effective therapeutic strategy in patients with solid tumors. Several trials have been conducted on immune-checkpoint inhibitor therapy in patients with malignant lymphoma and their efficacy has been reported. For example, in Hodgkin lymphoma, immune-checkpoint blockade has resulted in response rates of 65% to 75%. However, in non-Hodgkin lymphoma, the response rate to immune-checkpoint blockade was lower. In this review, we evaluate the biology of immune-checkpoint inhibition and the current data on its efficacy in malignant lymphoma, and identify the cases in which the treatment was more effective. -

Targeting Costimulatory Molecules in Autoimmune Disease
Targeting costimulatory molecules in autoimmune disease Natalie M. Edner1, Gianluca Carlesso2, James S. Rush3 and Lucy S.K. Walker1 1Institute of Immunity & Transplantation, Division of Infection & Immunity, University College London, Royal Free Campus, London, UK NW3 2PF 2Early Oncology Discovery, Early Oncology R&D, AstraZeneca, Gaithersburg, MD, USA 3Autoimmunity, Transplantation and Inflammation Disease Area, Novartis Institutes for Biomedical Research, Basel, Switzerland *Correspondence: Professor Lucy S.K. Walker. Institute of Immunity & Transplantation, Division of Infection & Immunity, University College London, Royal Free Campus, London, UK NW3 2PF. Tel: +44 (0)20 7794 0500 ext 22468. Email: [email protected]. 1 Abstract Therapeutic targeting of immune checkpoints has garnered significant attention in the area of cancer immunotherapy, and efforts have focused in particular on the CD28 family members CTLA-4 and PD-1. In autoimmunity, these same pathways can be targeted to opposite effect, to curb the over- exuberant immune response. The CTLA-4 checkpoint serves as an exemplar, whereby CTLA-4 activity is blocked by antibodies in cancer immunotherapy and augmented by the provision of soluble CTLA-4 in autoimmunity. Here we review the targeting of costimulatory molecules in autoimmune disease, focusing in particular on the CD28 family and TNFR family members. We present the state-of-the-art in costimulatory blockade approaches, including rational combinations of immune inhibitory agents, and discuss the future opportunities and challenges in this field. 2 The risk of autoimmune disease is an inescapable consequence of the manner in which the adaptive immune system operates. To ensure effective immunity against a diverse array of unknown pathogens, antigen recognition systems based on random gene rearrangement and mutagenesis have evolved to anticipate the antigenic universe. -

Pancancer IO360 Human Vapril2018
Gene Name Official Full Gene name Alias/Prev Symbols Previous Name(s) Alias Symbol(s) Alias Name(s) A2M alpha-2-macroglobulin FWP007,S863-7,CPAMD5 ABCF1 ATP binding cassette subfamily F member 1 ABC50 ATP-binding cassette, sub-family F (GCN20),EST123147 member 1 ACVR1C activin A receptor type 1C activin A receptor, type IC ALK7,ACVRLK7 ADAM12 ADAM metallopeptidase domain 12 a disintegrin and metalloproteinase domainMCMPMltna,MLTN 12 (meltrin alpha) meltrin alpha ADGRE1 adhesion G protein-coupled receptor E1 TM7LN3,EMR1 egf-like module containing, mucin-like, hormone receptor-like sequence 1,egf-like module containing, mucin-like, hormone receptor-like 1 ADM adrenomedullin AM ADORA2A adenosine A2a receptor ADORA2 RDC8 AKT1 AKT serine/threonine kinase 1 v-akt murine thymoma viral oncogene homologRAC,PKB,PRKBA,AKT 1 ALDOA aldolase, fructose-bisphosphate A aldolase A, fructose-bisphosphate ALDOC aldolase, fructose-bisphosphate C aldolase C, fructose-bisphosphate ANGPT1 angiopoietin 1 KIAA0003,Ang1 ANGPT2 angiopoietin 2 Ang2 ANGPTL4 angiopoietin like 4 angiopoietin-like 4 pp1158,PGAR,ARP4,HFARP,FIAF,NL2fasting-induced adipose factor,hepatic angiopoietin-related protein,PPARG angiopoietin related protein,hepatic fibrinogen/angiopoietin-related protein,peroxisome proliferator-activated receptor (PPAR) gamma induced angiopoietin-related protein,angiopoietin-related protein 4 ANLN anillin actin binding protein anillin (Drosophila Scraps homolog), actin bindingANILLIN,Scraps,scra protein,anillin, actin binding protein (scraps homolog, Drosophila) -

The Indoleamine 2,3 Dioxygenase Pathway Drives Intratumoral B Cell
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.25.456776; this version posted August 27, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The indoleamine 2,3 dioxygenase pathway drives intratumoral B cell maintenance. by Burles A. Johnson III1,2,3, Adam K. Aragaki2, Donna M. Williams3, Ophelia Rogers3, Jack Mountain2, Li Luo3, Wenhao Zhang3, Lingling Xian3, Mingxiao Feng2, Lionel Chia3, Dominic Dordai4, Noah M. Hahn1,2, Stephen Desiderio4, Theodore S. Johnson5, David J. McConkey2, and Linda M.S. Resar1,3,6. 1Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, MD 21205 2Johns Hopkins Greenberg Bladder Cancer Institute, Johns Hopkins University School of Medicine, 600 North Wolfe Street, Park 219, Baltimore, MD 21287 3Division of Hematology, Department of Medicine, Johns Hopkins University School of Medicine, 720 Rutland Avenue, Ross Research Building, Room 1025, Baltimore, MD 21205 4Institute for Basic Biomedical Sciences, Johns Hopkins University School of Medicine, Baltimore, MD 21205 5Georgia Cancer Center and Department of Pediatrics, Augusta University, 1120 15th Street, Augusta, GA 30912 6Department of Pathology and Institute for Cellular Engineering, Johns Hopkins University School of Medicine, Baltimore, MD 21205 Abstract: 219 words; Manuscript Text: 4929 words Correspondence: [email protected] or [email protected] Abbreviations: IDO1, indoleamine 2,3 dioxygenase-1; Bregs, regulatory B cells, Tregs, regulatory T cells; LLC, Lewis Lung Carcinoma; PD-1/L, programmed cell death protein-1/ligand; APCs, antigen presenting cells; MDSCs, myeloid derived suppressor cells; IL, interleukin. bioRxiv preprint doi: https://doi.org/10.1101/2021.08.25.456776; this version posted August 27, 2021. -

Methylation Epigenetic Landscape of Cpg DNA Defining CD4 T Cell Memory By
The Journal of Immunology Defining CD4 T Cell Memory by the Epigenetic Landscape of CpG DNA Methylation H. Kiyomi Komori, Traver Hart, Sarah A. LaMere, Pamela V. Chew, and Daniel R. Salomon Memory T cells are primed for rapid responses to Ag; however, the molecular mechanisms responsible for priming remain incom- pletely defined. CpG methylation in promoters is an epigenetic modification, which regulates gene transcription. Using targeted bisulfite sequencing, we examined methylation of 2100 genes (56,000 CpGs) mapped by deep sequencing of T cell activation in human naive and memory CD4 T cells. Four hundred sixty-six CpGs (132 genes) displayed differential methylation between naive and memory cells. Twenty-one genes exhibited both differential methylation and gene expression before activation, linking pro- moter DNA methylation states to gene regulation; 6 of 21 genes encode proteins closely studied in T cells, whereas 15 genes represent novel targets for further study. Eighty-four genes demonstrated differential methylation between memory and naive cells that cor- related to differential gene expression following activation, of which 39 exhibited reduced methylation in memory cells coupled with increased gene expression upon activation compared with naive cells. These reveal a class of primed genes more rapidly expressed in memory compared with naive cells and putatively regulated by DNA methylation. These findings define a DNA methylation sig- nature unique to memory CD4 T cells that correlates with activation-induced gene expression. The Journal of Immunology, 2015, 194: 1565–1579. ifferentiation into fast-acting memory cells following Ag have a demethylated IL-4 gene promoter (10), do not express IFN-g, recognition is a central tenet of T cell immunology. -

A New Horizon for the Treatment of Multiple Myeloma
review Investigational agents in immunotherapy: a new horizon for the treatment of multiple myeloma Cindy Varga,1 Jacob P. Laubach,2 Kenneth C. Anderson2 and Paul G. Richardson2 1Tufts Medical Center, Division of Hematology-Oncology, Boston, MA and 2Dana Farber Cancer Institute, Harvard Medical School, Jerome Lipper Multiple Myeloma Center, Boston, MA, USA Summary experts must rely on other anti-plasma cell approaches, which include alkylator agents (cyclophosphamide, oral mel- The treatment of multiple myeloma (MM) has gone through phalan), immunomodulators (IMiDs) (thalidomide, lenalido- several major advances over the last 5 years with the intro- mide, pomalidomide) and proteasome inhibitors (PIs) duction of next generation proteasome inhibitors (PI; carfil- (bortezomib, carfilzomib, ixazomib). Triplet combinations of zomib, ixazomib) and immunomodulatory derivatives these agents have proven to be quite effective with manage- (IMiD; pomalidomide), with these new agents having a sub- able toxicity profiles overall. Over the last few years, the stantial impact on patient outcome. However, despite these treatment algorithm for MM has undergone a major over- advances, MM remains a highly resistant disease given its haul with the introduction of humanized monoclonal anti- propensity for clonal heterogeneity and its complex interac- bodies (mAbs). Daratumumab and elotuzumab are mAbs tion with the surrounding bone marrow microenvironment. that effectively target CD38 and signalling lymphocytic acti- Almost all patients eventually relapse despite therapeutic vation family 7 (SLAMF7) on the surface of plasma cells, responses to a PI, IMiD or both. With the regulatory respectively, and have been approved by the US Food and approval of the monoclonal antibodies Daratumumab and Drug Administration (FDA) for the treatment of relapsed Elotuzumab in 2015, impressive and durable responses are refractory disease as of 2015. -

Mouse CD Marker Chart Bdbiosciences.Com/Cdmarkers
BD Mouse CD Marker Chart bdbiosciences.com/cdmarkers 23-12400-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1d CD1.1, CD1.2, Ly-38 Lipid, Glycolipid Ag + + + + + + + + CD104 Integrin b4 Laminin, Plectin + DNAX accessory molecule 1 (DNAM-1), Platelet and T cell CD226 activation antigen 1 (PTA-1), T lineage-specific activation antigen 1 CD112, CD155, LFA-1 + + + + + – + – – CD2 LFA-2, Ly-37, Ly37 CD48, CD58, CD59, CD15 + + + + + CD105 Endoglin TGF-b + + antigen (TLiSA1) Mucin 1 (MUC1, MUC-1), DF3 antigen, H23 antigen, PUM, PEM, CD227 CD54, CD169, Selectins; Grb2, β-Catenin, GSK-3β CD3g CD3g, CD3 g chain, T3g TCR complex + CD106 VCAM-1 VLA-4 + + EMA, Tumor-associated mucin, Episialin + + + + + + Melanotransferrin (MT, MTF1), p97 Melanoma antigen CD3d CD3d, CD3 d chain, T3d TCR complex + CD107a LAMP-1 Collagen, Laminin, Fibronectin + + + CD228 Iron, Plasminogen, pro-UPA (p97, MAP97), Mfi2, gp95 + + CD3e CD3e, CD3 e chain, CD3, T3e TCR complex + + CD107b LAMP-2, LGP-96, LAMP-B + + Lymphocyte antigen 9 (Ly9), -
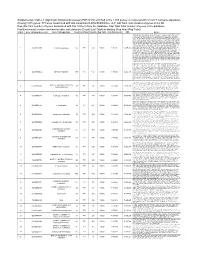
(FDR<0.05) Enriched in the 1015 Genes Co-Clustered with Known T
Supplementary Table 3. Significant biological processes (FDR<0.05) enriched in the 1,015 genes co-clustered with known T cell gene signatures. Among 1,015 genes, 771 were associated with GO annotation in DAVID database v6.7. List Total: total number of genes in my list. Pop Hits: total number of genes associated with this GO term from the database. Pop Total: total number of genes in the database. Fold Enrichment: relative enrichment ratio, calculated by (Count)/(List Total) divided by (Pop Hits)/(Pop Total). Index Gene Ontology Accession Gene Ontology Name Count List Total Pop Hits Pop Total Fold Enrichment FDR Genes AQP9, C1QC, B2M, LILRA1, LILRA2, CLEC4E, S1PR4, LILRA4, IFNG, LILRA6, CLEC4A, VNN1, ERAP2, FAS, CRTAM, C5AR1, GBP5, NCF2, NCF1, NCF4, SERPING1, HLA-DQA2, HLA-DQA1, PDCD1LG2, LILRB1, CCR9, C1QA, C1QB, LILRB2, CCR7, CCR6, UNC13D, CCR5, CD40LG, CCR4, LILRB3, CCR2, LILRB4, HLA-DPA1, VSIG4, HLA-DRA, IL1R2, IL1R1, HLA-DRB1, OAS3, ACP5, OAS1, OAS2, CD74, IFI35, ZAP70, FCER1G, HLA-DRB5, HLA-DPB1, HLA-DOA, HLA-DOB, DHX58, BLNK, IL23R, KIR2DS4, CD300C, SLAMF7, OASL, RGS1, APOL1, CD300A, HMHB1, CD209, CLEC7A, LY86, LY9, CLNK, FCRL4, SH2D1A, NOD2, HAMP, CCL3L1, CCL3L3, TICAM2, ICAM1, GZMA, CMKLR1, LY96, WAS, IL18BP, LAX1, TNFSF12- TNFSF13, HLA-DQB1, CSF2, GPR183, CCR1, GPR65, CXCL9, NCF1C, IL7R, CLEC10A, CCL24, CCL22, CYP27B1, CCL23, FCGR1C, FTHL3, FCGR1A, FCGR1B, BCL3, C2, CD27, CD28, FYB, IL18R1, IL7, CD1C, CTLA4, CCL19, CD1B, CD1A, TRIM22, CD180, CD1E, CCL18, CCL17, CCL13, FCGR2B, FCGR2C, P2RY14, LIME1, CD14, IL16, IL18, TLR1, TNFSF15, -

Gene Expression Targets
Supplementary Table S1: Gene expression targets Cell Functional Probe ID Gene ID Gene Descriptions type Category Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19 molecule B 1 MmugDNA.14306.1.S1_at CD1C CD1c molecule B 1 MmugDNA.20774.1.S1_at CD274 CD274 molecule B 1 MmugDNA.19231.1.S1_at CD28 CD28 molecule B 1 MmuSTS.3490.1.S1_at CD72 CD72 molecule B 1 MmugDNA.37827.1.S1_at CD79B CD79b molecule, immunoglobulin- associated beta B 1 MmugDNA.29916.1.S1_at CD80 CD80 molecule B 1 MmugDNA.6430.1.S1_at CD83 CD83 molecule B 1 MmugDNA.32699.1.S1_at CD86 CD86 molecule B 1 MmugDNA.19832.1.S1_at CIITA class II, major histocompatibility complex, transactivator B 1 MmuSTS.3942.1.S1_at CR2 complement component receptor 2 B 1 MmugDNA.25496.1.S1_at CTLA4 cytotoxic T-lymphocyte-associated protein 4 B 1 MmugDNA.5901.1.S1_at FAS Fas cell surface death receptor B 1 MmugDNA.8857.1.S1_at FASLG Fas ligand (TNF superfamily, member 6) B 1 MmugDNA.27012.1.S1_at FKBP2 FK506 binding protein 2, 13kDa P 1 MmugDNA.38565.1.S1_at FOS FBJ murine osteosarcoma viral oncogene homolog B 1 MmuSTS.165.1.S1_at ID3 inhibitor of DNA binding 3 B 1 MmugDNA.20492.1.S1_at IGBP1 immunoglobulin (CD79A) binding protein 1 B 1 MmugDNA.2488.1.S1_at IGHD immunoglobulin heavy constant delta B 1 MmuSTS.4350.1.S1_at IGHM immunoglobulin heavy constant mu B 1 MmunewRS.438.1.S1_at IL2 interleukin 2 B 1 MmugDNA.42882.1.S1_x_at SPN sialophorin B,P 1 MmuSTS.4526.1.S1_at SYK spleen tyrosine kinase B 1 MmuSTS.4672.1.S1_at TNF tumor necrosis factor -

Prevalence of PDL1 Amplification and Preliminary Response to Immune Checkpoint Blockade in Solid Tumors
Supplementary Online Content Goodman AM, Piccioni D, Kato S, et al. Prevalence of PDL1 Amplification and Preliminary Response to Immune Checkpoint Blockade in Solid Tumors. JAMA Oncol. Published online June 14, 2018. doi:10.1001/jamaoncol.2018.1701 Supplement. eFigure. CONSORT Diagram eTable 1. Biomarkers for Response to Immunotherapy in 13 Patients With PD-L1 Amplification Seen at University of California, San Diego Moores Cancer Center eTable 2. Definitions for PD-L1 IHC Positivity eTable 3. PDL1 Amplification by Solid Tumor Type (PDL1 Amplification Was Seen in 0.7% of 118 187 Samples With Malignancies) eTable 4. Clinical Characteristics of 13 Patients With PDL1 and/or PDCD1LG2 (PDL2) Amplifications Seen at UCSD Center for Personalized Cancer Therapy eTable 5. Patient Profiles and treatment Response eTable 6. PDL1/L2 Alterations Other Than Amplifications eFigure 2. Patient # 4 with glioblastoma This supplementary material has been provided by the authors to give readers additional information about their work. © 2018 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/28/2021 0.7% samples with 118,187 samples at Foundation PD-L1 CNAs Medicine 2,039 samples at UC San Diego Center for Personalized Cancer Therapy 13 patients identified with PD-L1 CNAs seen at UCSD 9 (69%) patients treated with 4 (31%) patients not treated with checkpoint blockade checkpoint blockade 1 (8%) patient treated with allogeneic stem cell transplantation eFigure. CONSORT Diagram Abbreviations: CNAs = copy number alterations; UC = University of California © 2018 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/28/2021 eTable 1. -

Genomic Amplification of CD274 (PD-L1) in Small-Cell Lung Cancer
Published OnlineFirst September 12, 2016; DOI: 10.1158/1078-0432.CCR-16-1069 Personalized Medicine and Imaging Clinical Cancer Research Genomic Amplification of CD274 (PD-L1) in Small-Cell Lung Cancer Julie George1, Motonobu Saito2,3, Koji Tsuta4, Reika Iwakawa2, Kouya Shiraishi2, Andreas H. Scheel5, Shinsuke Uchida4, Shun-ichi Watanabe6, Ryo Nishikawa7, Masayuki Noguchi8, Martin Peifer9, Se Jin Jang10, Iver Petersen11, Reinhard Buttner€ 5, Curtis C. Harris12, Jun Yokota2,13, Roman K. Thomas1,5,14, and Takashi Kohno2,15 Abstract Purpose: Programmed death ligand-1 (PD-L1), encoded by was further examined by transcriptome sequencing and immu- the CD274 gene, is a target for immune checkpoint blockade; nohistochemical (IHC) staining. however, little is known about genomic CD274 alterations. A Results: By examining somatic copy number alterations in two subset of small-cell lung cancer (SCLC) exhibits increased copy cohorts of primary human SCLC specimens, we observed 9p24 number of chromosome 9p24, on which CD274 resides; how- copy number gains (where CD274 resides) and focal, high-level ever, most SCLCs show low expression of PD-L1. We therefore amplification of CD274. We found evidence for genomic targeting examined whether CD274 is a target of recurrent genomic of CD274, suggesting selection during oncogenic transformation. alterations. CD274 amplification was caused by genomic rearrangements not Experimental Design: We examined somatic copy number affecting the open reading frame, thus leading to massively alterations in two patient cohorts by quantitative real-time PCR increased CD274 transcripts and high level expression of PD-L1. in 72 human SCLC cases (cohort 1) and SNP array analysis in Conclusions: A subset (4/210, 1.9%) of human SCLC patient 138 human SCLC cases (cohort 2).