Copyrighted Material
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Study of the Partial and Exhaustive Intramolecular Dehydration of D-Sorbitol
Available online at http://journal-of-agroalimentary.ro Journal of Journal of Agroalimentary Processes and Agroalimentary Processes and Technologies 2013, 19(2), 259-270 Technologies Study of the partial and exhaustive intramolecular dehydration of D-sorbitol Ileana Cocan *, Monica Viorica Negrea, Ionel Vasile Jianu ”Dunărea de Jos” University of Galati, Faculty of Food Science and Engineering, Domnească Street, 47, RO-800008, Galati, Romania Received: 07 February 2013; Accepted: 09 March 2013 ______________________________________________________________________________________ Abstract Sweetener, humectants, sequestrant, texturizer, stabilizer, bulking agent, D-sorbitol represents a leading product in the processing of the polysorbate class. The scope of this work is to obtain details on the preparation and accession of D-sorbitol mono- and dianhydride as a method of chemical protection of primary and secondary hydroxyl functional groups (1:4) (3:6) as part of the strategy of controlled processing of polysorbates containing “homogeneous” polyoxyethylenic chains (n = 3, 6, 9, 18). This work is investigating the dependence of yield of internal dehydration (partial and/or exhaustive, direct of D-sorbitol) on temperature (100-160°C and 120-200°C , respectively ) and duration (5–35 minutes and 10–100 minutes , respectively ) for the processing of 1,4-sorbitan and 2,5-isosorbide. The evolution of the hydroxyl number (mg KOH/g D-sorbitol) (mg KOH/g 1,4-sorbitan) related to the theoretical (initial) value was followed and optimal values for the yield and dehydration parameters were established. Also in this work, the mathematical modeling of the dependence curves experimentally registered is realized. Keywords : izosorbide, D-sorbitol, D-glucitol, D-sorbit, 1,4-sorbitan ______________________________________________________________________________________ 1. -

Stereochemistry CHAPTER3
28 Stereochemistry CHAPTER3 Stereochemistry Stereochemistry : It involves the study of the relative spatial arrangement of atoms within the molecules. Dynamic stereochemistry: Dynamic stereochemistry is the study of the effect of stereochemistry on the rate of a chemical reaction. First Stereochemist – Louis Pasteur (1849): Significance of stereochemistry: One of the most infamous demonstration of the significance of stere- ochemistry was the Thalidomide disaster. Thalidomide is a drug was first prepared in 1957 in Germany, prescribed for treating morning Sickness in pregnant women. It was discovered that one optical isomer i.e. R- Isomer of the drug was safe whereas the S-isomer had teratogenic effect, causing serious genetic damage to early embryonic growth and development. O O H O O H N N 1 2 N 3 O N O H H 4 O R-isomer O S-isomer Drug for morning sickness in pregnant women. Teratogenic effect [Remark: In human body, Thalidomide undergoes racemization: even if only one of the two stereoisomers is ingested, the other one is produced.] Now we have another example - Propanolol. H H N N O O H OH OH H R-Propanolol (contraceptive) S-Propanolol (antihypertensive) Stereochemistry 29 SOME TERMINOLOGY Optical activity: The term optical activity derived from the interaction of chiral materials with polarized light. Scalemic: Any non-racemic chiral substance is called Scalemic. • A chiral substance is enantio pure or homochiral when only one of two possible enantiomer is present. • A chiral substance is enantio enriched or heterochiral when an excess of one enantiomer is present but not the exclusion of the other. -

Abstract Multicomponent Reactions of Salicylaldehyde, Cyclic Ketones, and Arylamines Through Cooperative Enamine-Metal Lewis
ABSTRACT MULTICOMPONENT REACTIONS OF SALICYLALDEHYDE, CYCLIC KETONES, AND ARYLAMINES THROUGH COOPERATIVE ENAMINE-METAL LEWIS ACID CATALYSIS by Ryan Gregory Sarkisian Multicomponent reactions (MCRs) are the most atom economic, highly selective, and convergent type of reaction. This allows for a reaction to have a wide scope and allows for maximization of the complexity of a product. Catalyzing these MCRs with asymmetric catalysis is a novel way to introduce stereocontrol into highly complex molecules with various functional groups. Asymmetric catalysis is considered the most efficient method for constructing highly functionalized optically active stereopure compounds. There are three pillars of asymmetric catalysis: biocatalysis, transition metal catalysis, and organocatalysis. This research focuses on two of these pillars, transition metal catalysis and organocatalysis, working cooperatively to catalyze this MCR. The focus is to educate or refresh the audience on the basic topics that make up the complexity of the MCRs being catalyzed by cooperative asymmetric catalysis. Ultimately to explore the cooperative catalysts used to synthesize both the racemic and asymmetric three fused ring products (9-((4-methoxyphenyl)amino)-2,3,4,4a,9,9a-hexahydro-1H-xanthen-4a-ol). MULTICOMPONENT REACTIONS OF SALICYLALDEHYDE, CYCLIC KETONES, AND ARYLAMINES THROUGH COOPERATIVE ENAMINE-METAL LEWIS ACID CATALYSIS A THESIS Submitted to the Faculty of Miami University in partial fulfillment of the requirements for the degree of Master of Science Department of -

Catalytic Direct Asymmetric Michael Reactions
ORGANIC LETTERS 2001 Catalytic Direct Asymmetric Michael Vol. 3, No. 23 Reactions: Taming Naked Aldehyde 3737-3740 Donors Juan M. Betancort and Carlos F. Barbas III* The Skaggs Institute for Chemical Biology and the Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037 [email protected] Received September 5, 2001 ABSTRACT Direct catalytic enantio- and diastereoselective Michael addition reactions of unmodified aldehydes to nitro olefins using (S)-2-(morpholinomethyl)- pyrrolidine as a catalyst are described. The reactions proceed in good yield (up to 96%) in a highly syn-selective manner (up to 98:2) with enantioselectivities approaching 80%. The resulting γ-formyl nitro compounds are readily converted to chiral, nonracemic 3,4-disubstituted pyrrolidines. The Michael reaction is generally regarded as one of the Typically, carbon nucleophiles that contain an active most efficient carbon-carbon bond forming reactions, and methylene center such as malonic acid esters, â-keto esters, studies concerning this reaction have played an important nitroalkanes, etc. have been studied in the Michael reaction. role in the development of modern synthetic organic Carbonyl compounds, and ketones in particular, have gener- chemistry.1 As the demand for optically active compounds ally only been used as donors following their preactivation has soared in recent years, much progress has been made in by conversion into a more reactive species such as enol or the development of asymmetric variants of this reaction, enamine equivalents.5,6 In these cases, additional synthetic providing for the preparation of Michael adducts with high enantiomeric purity.2 Though remarkable advances have been (3) (a) Chataigner, I.; Gennari, C.; Ongeri, S.; Piarulli, U.; Ceccarelli, S. -

Organic Chemistry Frontiers
ORGANIC CHEMISTRY FRONTIERS View Article Online REVIEW View Journal | View Issue Progress in the synthesis of perylene bisimide dyes Cite this: Org. Chem. Front., 2019, 6, Agnieszka Nowak-Król and Frank Würthner * 1272 With their versatile absorption, fluorescence, n-type semiconducting and (photo-)stability properties, per- ylene bisimides have evolved as the most investigated compounds among polycyclic aromatic hydro- carbons during the last decade. In this review we collect the results from about 200 original publications, reporting a plethora of new perylene bisimide derivatives whose properties widely enrich the possibility for the application of these dyes beyond traditional fields. While some applications are highlighted, different from other recent reviews, our focus here is on the advances in the synthetic methodologies Received 17th December 2018, that have afforded new bay functionalizations, recently addressed functionalizations at the ortho-positions Accepted 17th February 2019 to the carbonyl groups, and annulation of carbo- and heterocyclic units. An impressive number of DOI: 10.1039/c8qo01368c perylene bisimide oligomers are highlighted as well which are connected by single bonds or spiro linkage rsc.li/frontiers-organic or in a fused manner, leading to arrays with fascinating optical and electronic properties. Creative Commons Attribution 3.0 Unported Licence. Introduction are the leading examples of the class of tetrapyrrole dyes, PBIs are the most important compounds of the family of polycyclic About a hundred years after their discovery, perylene-3,4:9,10- aromatic hydrocarbons. This outstanding role of PBIs has bis(dicarboximide)s, commonly abbreviated as PDIs or PBIs, evolved not only due to their properties, but also due to the have emerged as one of the most important classes of func- incredible development of the synthetic chemistry of this class tional dyes. -

Sorbitol Malabsorption 1/2
Internal Medicine and Gastroenterology Clinic Dres. Mares,M.Hanig,Mares, Hanig, Rambow, S.Blau, Blau, M.Seip, Hanig, Seip, A.Borchers,Blau,Kirchner, Hochstr. Hochstr. Hochstr. 43, 60313 43, 43, 60313 60313 Frankfurt/Main, Frankfurt/M., Frankfurt/M., www.gastroenterologie-ffm.de www.gastroenterologie-ffm.de www.gastroenterologie-ffm.de Nutrition hints for sorbitol malabsorption 1/2 Sorbitol intolerance Sorbitol incompatibility 1. What does this mean for nutrition? Avoid sorbitol as a sweetener and foods high in sorbitol. Small amount of sorbitol can often be tolerated (at most 10-10 g per day, sometimes less). In order to prevent general upper GI problems, easily digestible foods that do not cause gas are recommended. 2. Key points about foods – Avoid sorbitol and foods containing sorbitol – easily digestible food that does not cause gas 3. vegetables and fruits low in fibre (see 5) 4. Choice of foods The following foods are high in sorbitol and not suitable: – Sorbitol as sweetener: e.g. Sionon, Flarom, diabetic sweetener – Dietetic foods produced with sorbitol: for example, diabetic marmalades, diabetic sweets, diabetic baked goods – Types of fruit which have a naturally high sorbitol content: Apples, pears, cherries, prunes, plums, dates fruits with seeds, such as mirabelles, apricots, nectarines, and all fruit syrups made from these types of fruits – Types of fruit which have a naturally low sorbitol content: Berry fruits such as strawberries, raspberries, blackberries, blueberries, currants, gooseberries, citrus fruits, bananas, pineapples, kiwis – Sorbitol as a coating for: sultanas, raisins, and dried fruit or candied fruit – Sorbitol in sweets: chewing gum, jelly babies, jelly fruits, candies, chocolate bars, filled wafers, chocolate, etc. -

Organic Stereoisomerism
Organic : Stereoisomerism Stereoisomerism Introduction: When two compounds have same structure(hence same name) but still they are not identical and some of their properties are different then it means they are stereoisomers. They are not superimposable with each other. They differ in the spatial relationship between their atoms or groups. See this example. CH3 CH3 CH3 H CC CC H H H CH3 cis(Z)-but-2-ene trans(E)-but-2-ene Both are but-2-ene, but they are not identical. Their melting points and boiling points are different. They evolve different heat energy when hydrogenated. So what is the difference between them ? Is it structure- wise ? The answer is NO, as their structures are same i.e but-2-ene. They differ with respect the relationship between some atoms or groups in space. In cis-but-2-ene, the –CH3 groups are on the same side of the double bond and so also the H atoms. However, in trans-but-2-ene, the –CH3 groups lie on opposite sides of the double bond and so also the H atoms. For that reason, the two are not superimposable with each other. If you carry the model of cis-but-2-ene and try to overlap with trans-but-2-ene for the purpose of matching of groups and atoms, what do you find ? Are the two superimposable ? When one –CH3 group matches, the other not. When one –H atom matches, the other not. Thus the structures are non-superimposble. This kind of stereoisomerism is called Geometrical Isomerism(now-a-days called E-Z isomersm), because the geometrical relationsip between groups are different. -

The Use of Spirocyclic Scaffolds in Drug Discovery ⇑ Yajun Zheng , Colin M
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Bioorganic & Medicinal Chemistry Letters 24 (2014) 3673–3682 Contents lists available at ScienceDirect Bioorganic & Medicinal Chemistry Letters journal homepage: www.elsevier.com/locate/bmcl BMCL Digest The use of spirocyclic scaffolds in drug discovery ⇑ Yajun Zheng , Colin M. Tice, Suresh B. Singh Vitae Pharmaceuticals Inc., 502 West Office Center Drive, Fort Washington, PA 19034, United States article info abstract Article history: Owing to their inherent three-dimensionality and structural novelty, spiro scaffolds have been increas- Received 15 April 2014 ingly utilized in drug discovery. In this brief review, we highlight selected examples from the primary Revised 17 June 2014 medicinal chemistry literature during the last three years to demonstrate the versatility of spiro scaffolds. Accepted 27 June 2014 With recent progress in synthetic methods providing access to spiro building blocks, spiro scaffolds are Available online 5 July 2014 likely to be used more frequently in drug discovery. Ó 2014 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND Keywords: license (http://creativecommons.org/licenses/by-nc-nd/3.0/). Spirocyclic Scaffold Drug discovery One widely used strategy in drug design is to rigidify the ligand present in a number of bioactive natural products such as mito- conformation by introducing a ring.1 The resulting cyclic analog mycins, it is generally difficult to construct such aziridines from will suffer a reduced conformational entropy penalty upon binding unfunctionalized olefins. A recent report of a facile synthetic route to a protein target. -

The Arene–Alkene Photocycloaddition
The arene–alkene photocycloaddition Ursula Streit and Christian G. Bochet* Review Open Access Address: Beilstein J. Org. Chem. 2011, 7, 525–542. Department of Chemistry, University of Fribourg, Chemin du Musée 9, doi:10.3762/bjoc.7.61 CH-1700 Fribourg, Switzerland Received: 07 January 2011 Email: Accepted: 23 March 2011 Ursula Streit - [email protected]; Christian G. Bochet* - Published: 28 April 2011 [email protected] This article is part of the Thematic Series "Photocycloadditions and * Corresponding author photorearrangements". Keywords: Guest Editor: A. G. Griesbeck benzene derivatives; cycloadditions; Diels–Alder; photochemistry © 2011 Streit and Bochet; licensee Beilstein-Institut. License and terms: see end of document. Abstract In the presence of an alkene, three different modes of photocycloaddition with benzene derivatives can occur; the [2 + 2] or ortho, the [3 + 2] or meta, and the [4 + 2] or para photocycloaddition. This short review aims to demonstrate the synthetic power of these photocycloadditions. Introduction Photocycloadditions occur in a variety of modes [1]. The best In the presence of an alkene, three different modes of photo- known representatives are undoubtedly the [2 + 2] photocyclo- cycloaddition with benzene derivatives can occur, viz. the addition, forming either cyclobutanes or four-membered hetero- [2 + 2] or ortho, the [3 + 2] or meta, and the [4 + 2] or para cycles (as in the Paternò–Büchi reaction), whilst excited-state photocycloaddition (Scheme 2). The descriptors ortho, meta [4 + 4] cycloadditions can also occur to afford cyclooctadiene and para only indicate the connectivity to the aromatic ring, and compounds. On the other hand, the well-known thermal [4 + 2] do not have any implication with regard to the reaction mecha- cycloaddition (Diels–Alder reaction) is only very rarely nism. -

Polyols Have a Variety of Functional Properties That Make Them Useful Alternatives to Sugars in Applications Including Baked Goods
Polyols have a variety of functional properties that make them useful alternatives to sugars in applications including baked goods. Photo © iStockphoto.com/Synergee pg 22 09.12 • www.ift.org BY LYN NABORS and THERESA HEDRICK SUGAR REDUCTION WITH Polyols Polyols are in a unique position to assist with reduced-sugar or sugar-free reformulations since they can reduce calories and complement sugar’s functionality. ugar reduction will be an important goal over the of the product’s original characteristics may still be main- next few years as consumers, government, and in- tained with the replacement of those sugars by polyols. Sdustry alike have expressed interest in lower-calorie In addition, excellent, good-tasting sugar-free products and lower-sugar foods. The 2010 Dietary Guidelines for can be developed by using polyols. Polyols are in a unique Americans put a strong emphasis on consuming fewer position to assist with reduced-sugar or sugar-free refor- calories and reducing intake of added sugars. The In- mulations; since they are only partially digested and ab- stitute of Medicine (IOM) held a public workshop in sorbed, they can reduce calories and complement sugar’s November 2010 to discuss ways the food industry can functionality. Polyols provide the same bulk as sugars and use contemporary and innovative food processing tech- other carbohydrates. Additionally, polyols have a clean, nologies to reduce calorie intake in an effort to reduce sweet taste, which is important since consumers are not and prevent obesity, and in October 2011 recommended likely to sacrifice taste for perceived health benefits. Poly- front-of-package labeling that includes rating the product ols have a host of other functional properties that make based on added sugars content. -

Comparative Total Syntheses of Strychnine
Comparative Total Syntheses of Strychnine N C D MacMillan Group Meeting N H O H A H B E Nathan Jui H N N H H F G July 22, 2009 O O O References: Pre-Volhardt: Bonjoch Chem. Rev. 2000, 3455. Woodward Tetrahedron, 1963, 247. Volhardt J. Am. Chem. Soc. 2001, 9324. Magnus J. Am. Chem. Soc. 1993, 8116. Martin J. Am. Chem. Soc. 2001, 8003. Overman J. Am. Chem. Soc. 1995, 5776. Bodwell Angew. Chem. Int. Ed. 2002, 3261. Kuehne J. Org. Chem. 1993, 7490. Mori J. Am. Chem. Soc. 2003, 9801. Kuehne J. Org. Chem. 1998, 9427. Shibasaki Tetrahedron 2004, 9569. Rawal J. Org. Chem. 1994, 2685. Fukuyama J. Am. Chem. Soc. 2004, 10246. Bosch, Bonjoch Chem. Eur. J. 2000, 655. Padwa Org. Lett. 2007, 279. History and Structure of (!)-Strychnine ! Isolated in pure form in 1818 (Pelletier and Caventou) ! Structural Determination in 1947 (Robinson and Leuchs) ! 24 skeletal atoms (C21H22N2O2) ! Isolated in pure form in 1818 (Pelletier and Caventou) N ! Over !25 07 -pruinbglisc,a 6ti-osntesr epoecrteaninteinrgs to structure ! Structural Determination in 1947 (Robinson and Leuchs) ! Notor!io u Ss ptoirxoicne (nletethr a(lC d-o7s) e ~10-50 mg / adult) N H H ! Over 250 publications pertaining to structure O O ! Comm!o nClyD uEs eridn gr osdyesntet mpoison ! Notorious poison (lethal dose ~10-50 mg / adult) ! Hydroxyethylidine Strychnos nux vomica ! $20.20 / 10 g (Aldrich), ~1.5 wt% (seeds), ~1% (blossoms) "For it's molecular size it is the most complex substance known." -Robert Robinson History and Structure of (!)-Strychnine ! 24 skeletal atoms (C21H22N2O2) N ! 7-rings, 6-stereocenters ! Spirocenter (C-7) N H H O O ! CDE ring system ! Hydroxyethylidine "For it's molecular size it is the most complex substance known." -Robert Robinson "If we can't make strychnine, we'll take strychnine" -R. -
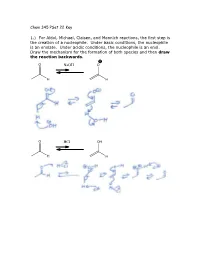
Chem 345 Pset 22 Key 1.) for Aldol, Michael, Claisen, and Mannich
Chem 345 PSet 22 Key 1.) For Aldol, Michael, Claisen, and Mannich reactions, the first step is the creation of a nucleophile. Under basic conditions, the nucleophile is an enolate. Under acidic conditions, the nucleophile is an enol. Draw the mechanism for the formation of both species and then draw the reaction backwards. O NaOH O H H O HCl OH H H 2.) In the case of a Mannich reaction, the electrophile (an imine) must also be made. Draw the mechanism for the forward and backward reaction: O MeNH2 N H H cat. AcOH H H 3.) When a secondary amine is used and the carbonyl has an enolizable proton, an enamine can be formed. Are enamines nucleophilic or electrophilic? Nucleophilic H O N N H2O H cat. AcOH H enamine N N H H enamine 4.) The following reaction is the Lobry-de-Bruyn-von-Ekenstein reaction. Don’t worry about the name. I only recently found out that that was the official name for the rearrangement leading from glucose to fructose or mannose and back again. I was taught it under a different name: the Carbohydrate Game. Under acidic or basic conditions, it is possible that the following sugars (glucose, fructose, and mannose) are in equilibrium with each other. Draw the acid catalyzed version and the base catalyzed version. (Hint: this is just a repeat of question 1.) O O H H H2C OH + + HO H H3O H OH H3O O or or HO- HO- HO H HO H HO H H OH H OH H OH H OH H OH H OH H2C OH H2C OH H2C OH D-glucose D-fructose D-mannose O O H H H2C OH + + HO H H3O H OH H3O O or or HO- HO- HO H HO H HO H H OH H OH H OH H OH H OH H OH H2C OH H2C OH H2C OH D-glucose D-fructose D-mannose The sugars are drawn in a Fisher projection (or what I think of is a dead cat projection).