Mediated Transgene Knockin at the H11 Locus in Pigs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Goat Anti-4E-T / EIF4ENIF1 Antibody Catalog No: Tcva06230
Web: www.taiclone.com Tel: +886-2-2735-9682 Email: [email protected] Goat anti-4E-T / EIF4ENIF1 Antibody Catalog No: tcva06230 Available Sizes Size: 100µg Specifications Application: Pep-ELISA, WB, IHC Research Area: RNA-sorting; mRNA decay; P-body; NIF transporter; translation Species Reactivity: Human, Mouse, Rat, Dog, Pig, Cow Host Species: Goat Immunogen / Amino acids: C-AKVISVDELEYRQ Conjugation: Unconjugated Form: Liquid Storage Buffer: Tris saline, 0.02% sodium azide, pH7.3 with 0.5% bovine serum albumin Concentration: 0.5 mg/ml in 200 µl Recommended Dilution: Western Blot: Approx 140-150kDa band observed in 293 lysates (predicted size of approx. 108kDa according to NP_062817.1 however our observation agrees with that of Dostie et al (see below) ). Recommended for use at 0.25-0.5µg/ml Copyright 2021 Taiclone Biotech Corp. Web: www.taiclone.com Tel: +886-2-2735-9682 Email: [email protected] Peptide ELISA: antibody detection limit dilution 1:32000. Amino Acid Sequence: NP_062817.2; NP_001157974.1 Storage Instruction: Aliquot store at -20C. Avoid freeze / thaw cycles. Alternative Names: EIF4ENIF1; 4E-T; Clast4; FLJ21601; 2610509L04Rik; eukaryotic translation initiation factor 4E nuclear import factor 1; eIF4E-transporter; FLJ26551; OTTHUMP00000063276 Gene ID: 56478 (human);74203 (mouse); Reference Sequence No.: NP_062817.2; NP_001157974.1 Calculated Molecular Weight: 108; 88.2 Purification: Purified from goat serum by ammonium sulphate precipitation followed by antigen affinity chromatography using the immunizing peptide DS Gene Ontology Terms: protein transporter; nucleocytoplasmic transport; cytoplasm; nucleus Positive Control: tcva06230p Notes This antibody is expected to recognise isoform 1 (NP_062817.2) and isoform b (NP_001157974.1). Reported variants represent identical protein (NP_062817.2; NP_001157973.1). -

Towards a Molecular Understanding of Microrna-Mediated Gene Silencing
REVIEWS NON-CODING RNA Towards a molecular understanding of microRNA-mediated gene silencing Stefanie Jonas and Elisa Izaurralde Abstract | MicroRNAs (miRNAs) are a conserved class of small non-coding RNAs that assemble with Argonaute proteins into miRNA-induced silencing complexes (miRISCs) to direct post-transcriptional silencing of complementary mRNA targets. Silencing is accomplished through a combination of translational repression and mRNA destabilization, with the latter contributing to most of the steady-state repression in animal cell cultures. Degradation of the mRNA target is initiated by deadenylation, which is followed by decapping and 5ʹ‑to‑3ʹ exonucleolytic decay. Recent work has enhanced our understanding of the mechanisms of silencing, making it possible to describe in molecular terms a continuum of direct interactions from miRNA target recognition to mRNA deadenylation, decapping and 5ʹ‑to‑3ʹ degradation. Furthermore, an intricate interplay between translational repression and mRNA degradation is emerging. Deadenylation MicroRNAs (miRNAs) are conserved post-transcriptional recruit additional protein partners to mediate silenc- 5,6 Shortening of mRNA poly(A) regulators of gene expression that are integral to ing . Silencing occurs through a combination of tails. In eukaryotes, this almost all known biological processes, including translational repression, deadenylation, decapping and process is catalysed by the cell growth, proliferation and differentiation, as well 5ʹ‑to‑3ʹ mRNA degradation5,6 (FIG. 1). The GW182 pro- consecutive but partially as organismal metabolism and development1. The teins play a central part in this process and are among redundant action of two 5,6 cytoplasmic deadenylase number of miRNAs encoded within the genomes of the most extensively studied AGO partners . -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Generation of H11-Albumin-Rtta Transgenic Mice: a Tool for Inducible Gene Expression in the Liver
INVESTIGATION Generation of H11-albumin-rtTA Transgenic Mice: A Tool for Inducible Gene Expression in the Liver Yu-Shan Li,*,1 Ran-Ran Meng,†,1 Xiu Chen,‡ Cui-Ling Shang,§ Hong-Bin Li,* Tao-Jun Zhang,† Hua-Yang Long,† Hui-Qi Li,† Yi-Jing Wang,† and Feng-Chao Wang†,2 *The School of Public Health, Xinxiang Medical University, Xinxiang, Henan, China 453003, †Transgenic Animal Center, National Institute of Biological Sciences, Beijing, China 102206, ‡Department of Pharmacy, Heze University, Heze, § Shandong, China, 274015, and Department of Reproductive Medicine, The Third Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan, China 453003 ABSTRACT The modification of the mouse genome by site-specific gene insertion of transgenes and other KEYWORDS genetic elements allows the study of gene function in different developmental stages and in the pathogenesis Hipp11 of diseases. Here, we generated a “genomic safe harbor” Hipp11 (H11)locus-specific knock-in transgenic albumin mouse line in which the albumin promoter is used to drive the expression of the reverse tetracycline promoter transactivator (rtTA) in the liver. The newly generated H11-albumin-rtTA transgenic mice were bred with doxycycline tetracycline-operator-Histone-2B-green fluorescent protein (TetO-H2BGFP) mice to assess inducibility and Tet-on system tissue-specificity. Expression of the H2BGFP fusion protein was observed exclusively upon doxycycline (Dox) induction in the liver of H11-albumin-rtTA/TetO-H2BGFP double transgenic mice. To further analyze the ability of the Dox-inducible H11-albumin-rtTA mice to implement conditional DNA recombination, H11- albumin-rtTA transgenic mice were crossed with TetO-Cre and Ai14 mice to generate H11-albumin-rtTA/ TetO-Cre/Ai14 triple transgenic mice. -

The Mechanism of Eukaryotic Translation Initiation and Principles of Its Regulation
REVIEWS POST-TRANSCRIPTIONAL CONTROL The mechanism of eukaryotic translation initiation and principles of its regulation Richard J. Jackson*, Christopher U. T. Hellen‡ and Tatyana V. Pestova‡ Abstract | Protein synthesis is principally regulated at the initiation stage (rather than during elongation or termination), allowing rapid, reversible and spatial control of gene expression. Progress over recent years in determining the structures and activities of initiation factors, and in mapping their interactions in ribosomal initiation complexes, have advanced our understanding of the complex translation initiation process. These developments have provided a solid foundation for studying the regulation of translation initiation by mechanisms that include the modulation of initiation factor activity (which affects almost all scanning-dependent initiation) and through sequence-specific RNA-binding proteins and microRNAs (which affect individual mRNAs). Met Translation initiation is the process of assembly of significance is particularly high, and we include evi- Met-tRNA i The unique initiator tRNA, elongation-competent 80S ribosomes, in which the ini- dence from lower eukaryotes only when it enhances our aminoacylated with tiation codon is base-paired with the anticodon loop understanding of the mechanisms in vertebrates. methionine, that is used to Met 1 of initiator tRNA (Met-tRNA i) in the ribosomal P-site . initiate protein synthesis. Mechanism of 5′ end-dependent initiation Its anticodon is complementary It requires at least nine eukaryotic initiation factors to the AUG initiation codon; (eIFs; TABLE 1) and comprises two steps: the formation The canonical mechanism of translation initiation can be it forms a specific ternary of 48S initiation complexes with established codon– divided into several stages (FIG.1), as described below. -

Mrna Export Through an Additional Cap-Binding Complex Consisting of NCBP1 and NCBP3
ARTICLE Received 2 Mar 2015 | Accepted 28 Jul 2015 | Published 18 Sep 2015 DOI: 10.1038/ncomms9192 OPEN mRNA export through an additional cap-binding complex consisting of NCBP1 and NCBP3 Anna Gebhardt1,*, Matthias Habjan1,*, Christian Benda2, Arno Meiler1, Darya A. Haas1, Marco Y. Hein3, Angelika Mann1, Matthias Mann3, Bianca Habermann4 & Andreas Pichlmair1 The flow of genetic information from DNA to protein requires polymerase-II-transcribed RNA characterized by the presence of a 50-cap. The cap-binding complex (CBC), consisting of the nuclear cap-binding protein (NCBP) 2 and its adaptor NCBP1, is believed to bind all capped RNA and to be necessary for its processing and intracellular localization. Here we show that NCBP1, but not NCBP2, is required for cell viability and poly(A) RNA export. We identify C17orf85 (here named NCBP3) as a cap-binding protein that together with NCBP1 forms an alternative CBC in higher eukaryotes. NCBP3 binds mRNA, associates with components of the mRNA processing machinery and contributes to poly(A) RNA export. Loss of NCBP3 can be compensated by NCBP2 under steady-state conditions. However, NCBP3 becomes pivotal under stress conditions, such as virus infection. We propose the existence of an alternative CBC involving NCBP1 and NCBP3 that plays a key role in mRNA biogenesis. 1 Innate Immunity Laboratory, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. 2 Department of Structural Cell Biology, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. 3 Department of Proteomics and Signal Transduction, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. 4 Bioinformatics Core Facility, Max-Planck Institute of Biochemistry, Martinsried, Munich D-82152, Germany. -

Systematic Discovery of Endogenous Human Ribonucleoprotein Complexes
bioRxiv preprint doi: https://doi.org/10.1101/480061; this version posted November 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Systematic discovery of endogenous human ribonucleoprotein complexes Anna L. Mallam1,2,3,§,*, Wisath Sae-Lee1,2,3, Jeffrey M. Schaub1,2,3, Fan Tu1,2,3, Anna Battenhouse1,2,3, Yu Jin Jang1, Jonghwan Kim1, Ilya J. Finkelstein1,2,3, Edward M. Marcotte1,2,3,§, Kevin Drew1,2,3,§,* 1 Department of Molecular Biosciences, University of Texas at Austin, Austin, TX 78712, USA 2 Center for Systems and Synthetic Biology, University of Texas at Austin, Austin, TX 78712, USA 3 Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, TX 78712, USA § Correspondence: [email protected] (A.L.M.), [email protected] (E.M.M.), [email protected] (K.D.) * These authors contributed equally to this work Short title: A resource of human ribonucleoprotein complexes Summary: An exploration of human protein complexes in the presence and absence of RNA reveals endogenous ribonucleoprotein complexes ! 1! bioRxiv preprint doi: https://doi.org/10.1101/480061; this version posted November 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Ribonucleoprotein (RNP) complexes are important for many cellular functions but their prevalence has not been systematically investigated. -

WO 2013/064702 A2 10 May 2013 (10.05.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2013/064702 A2 10 May 2013 (10.05.2013) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C12Q 1/68 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (21) International Application Number: HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, PCT/EP2012/071868 KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, (22) International Filing Date: ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, 5 November 20 12 (05 .11.20 12) NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, (25) Filing Language: English TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, (26) Publication Language: English ZM, ZW. (30) Priority Data: (84) Designated States (unless otherwise indicated, for every 1118985.9 3 November 201 1 (03. 11.201 1) GB kind of regional protection available): ARIPO (BW, GH, 13/339,63 1 29 December 201 1 (29. 12.201 1) US GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, (71) Applicant: DIAGENIC ASA [NO/NO]; Grenseveien 92, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, N-0663 Oslo (NO). -

Genetic Determination of the Ovarian Reserve: a Literature Review Aleksandra V
Moiseeva et al. J Ovarian Res (2021) 14:102 https://doi.org/10.1186/s13048-021-00850-9 REVIEW Open Access Genetic determination of the ovarian reserve: a literature review Aleksandra V. Moiseeva1, Varvara A. Kudryavtseva1, Vladimir N. Nikolenko1,2, Marine M. Gevorgyan3, Ara L. Unanyan1, Anastassia A. Bakhmet1 and Mikhail Y. Sinelnikov1,4* Abstract The ovarian reserve is one of the most important indicators of female fertility. It allows for the evaluation of the num- ber of viable oocytes. This parameter is actively used in pregnancy planning and in assisted reproductive technology application, as it determines chances of successful fertilization and healthy pregnancy. Due to increased attention towards diagnostic tests evaluating the ovarian reserve, there has been a growing interest in factors that infuence the state of the ovarian reserve. True reasons for pathological changes in the ovarian reserve and volume have not yet been explored in depth, and current diagnostic screening methods often fall short in efcacy. In the following review we analyze existing data relating to the study of the ovarian reserve through genetic testing, determining specifc characteristics of the ovarian reserve through genetic profling. We explore existing studies dedicated to fnding spe- cifc genetic targets infuencing the state of the ovarian reserve. Introduction ovarian hyperstimulation during in-vitro fertilization “Ovarian reserve” is a term that is used to describe the (IVF) treatment. However, many genetically determined remaining capacity of oocytes in the ovary. With age, the characteristics are not sufciently explored and require ovarian reserve tends to naturally diminish and normally further evaluation. In this review we aim to defne spe- does not present with pathological changes. -

Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations Fall 2010 Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress Renuka Nayak University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Computational Biology Commons, and the Genomics Commons Recommended Citation Nayak, Renuka, "Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress" (2010). Publicly Accessible Penn Dissertations. 1559. https://repository.upenn.edu/edissertations/1559 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1559 For more information, please contact [email protected]. Coexpression Networks Based on Natural Variation in Human Gene Expression at Baseline and Under Stress Abstract Genes interact in networks to orchestrate cellular processes. Here, we used coexpression networks based on natural variation in gene expression to study the functions and interactions of human genes. We asked how these networks change in response to stress. First, we studied human coexpression networks at baseline. We constructed networks by identifying correlations in expression levels of 8.9 million gene pairs in immortalized B cells from 295 individuals comprising three independent samples. The resulting networks allowed us to infer interactions between biological processes. We used the network to predict the functions of poorly-characterized human genes, and provided some experimental support. Examining genes implicated in disease, we found that IFIH1, a diabetes susceptibility gene, interacts with YES1, which affects glucose transport. Genes predisposing to the same diseases are clustered non-randomly in the network, suggesting that the network may be used to identify candidate genes that influence disease susceptibility. -
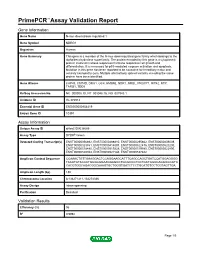
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name N-myc downstream regulated 1 Gene Symbol NDRG1 Organism Human Gene Summary This gene is a member of the N-myc downregulated gene family which belongs to the alpha/beta hydrolase superfamily. The protein encoded by this gene is a cytoplasmic protein involved in stress responses hormone responses cell growth and differentiation. It is necessary for p53-mediated caspase activation and apoptosis. Mutation in this gene has been reported to be causative for hereditary motor and sensory neuropathy-Lom. Multiple alternatively spliced variants encoding the same protein have been identified. Gene Aliases CAP43, CMT4D, DRG1, GC4, HMSNL, NDR1, NMSL, PROXY1, RIT42, RTP, TARG1, TDD5 RefSeq Accession No. NC_000008.10, NT_008046.16, NG_007943.1 UniGene ID Hs.372914 Ensembl Gene ID ENSG00000104419 Entrez Gene ID 10397 Assay Information Unique Assay ID qHsaCID0038665 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000356692, ENST00000488810, ENST00000295862, ENST00000409039, ENST00000323851, ENST00000414097, ENST00000522476, ENST00000520230, ENST00000518480, ENST00000519228, ENST00000519580, ENST00000522890, ENST00000520943, ENST00000521544, ENST00000537882 Amplicon Context Sequence CCAAACTGTTGAAGGACTCCAGGAAGCATTTCAGCCAGCTGATCCATGGAGGGG TACATGTACCCTGCGGGGAAGGAGGCTGCGCCGTCCTGCTGGCCAGGGGCGTC CACGTGGCAGACGGCAAAGTGCTGGGTGATCTCCTGCATGTCCTCGTAGTTGA Amplicon Length (bp) 130 Chromosome Location 8:134271411-134274386 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein