Genetics of Premature Ovarian Insufficiency and the Association with X-Autosome Translocations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Goat Anti-4E-T / EIF4ENIF1 Antibody Catalog No: Tcva06230
Web: www.taiclone.com Tel: +886-2-2735-9682 Email: [email protected] Goat anti-4E-T / EIF4ENIF1 Antibody Catalog No: tcva06230 Available Sizes Size: 100µg Specifications Application: Pep-ELISA, WB, IHC Research Area: RNA-sorting; mRNA decay; P-body; NIF transporter; translation Species Reactivity: Human, Mouse, Rat, Dog, Pig, Cow Host Species: Goat Immunogen / Amino acids: C-AKVISVDELEYRQ Conjugation: Unconjugated Form: Liquid Storage Buffer: Tris saline, 0.02% sodium azide, pH7.3 with 0.5% bovine serum albumin Concentration: 0.5 mg/ml in 200 µl Recommended Dilution: Western Blot: Approx 140-150kDa band observed in 293 lysates (predicted size of approx. 108kDa according to NP_062817.1 however our observation agrees with that of Dostie et al (see below) ). Recommended for use at 0.25-0.5µg/ml Copyright 2021 Taiclone Biotech Corp. Web: www.taiclone.com Tel: +886-2-2735-9682 Email: [email protected] Peptide ELISA: antibody detection limit dilution 1:32000. Amino Acid Sequence: NP_062817.2; NP_001157974.1 Storage Instruction: Aliquot store at -20C. Avoid freeze / thaw cycles. Alternative Names: EIF4ENIF1; 4E-T; Clast4; FLJ21601; 2610509L04Rik; eukaryotic translation initiation factor 4E nuclear import factor 1; eIF4E-transporter; FLJ26551; OTTHUMP00000063276 Gene ID: 56478 (human);74203 (mouse); Reference Sequence No.: NP_062817.2; NP_001157974.1 Calculated Molecular Weight: 108; 88.2 Purification: Purified from goat serum by ammonium sulphate precipitation followed by antigen affinity chromatography using the immunizing peptide DS Gene Ontology Terms: protein transporter; nucleocytoplasmic transport; cytoplasm; nucleus Positive Control: tcva06230p Notes This antibody is expected to recognise isoform 1 (NP_062817.2) and isoform b (NP_001157974.1). Reported variants represent identical protein (NP_062817.2; NP_001157973.1). -

Towards a Molecular Understanding of Microrna-Mediated Gene Silencing
REVIEWS NON-CODING RNA Towards a molecular understanding of microRNA-mediated gene silencing Stefanie Jonas and Elisa Izaurralde Abstract | MicroRNAs (miRNAs) are a conserved class of small non-coding RNAs that assemble with Argonaute proteins into miRNA-induced silencing complexes (miRISCs) to direct post-transcriptional silencing of complementary mRNA targets. Silencing is accomplished through a combination of translational repression and mRNA destabilization, with the latter contributing to most of the steady-state repression in animal cell cultures. Degradation of the mRNA target is initiated by deadenylation, which is followed by decapping and 5ʹ‑to‑3ʹ exonucleolytic decay. Recent work has enhanced our understanding of the mechanisms of silencing, making it possible to describe in molecular terms a continuum of direct interactions from miRNA target recognition to mRNA deadenylation, decapping and 5ʹ‑to‑3ʹ degradation. Furthermore, an intricate interplay between translational repression and mRNA degradation is emerging. Deadenylation MicroRNAs (miRNAs) are conserved post-transcriptional recruit additional protein partners to mediate silenc- 5,6 Shortening of mRNA poly(A) regulators of gene expression that are integral to ing . Silencing occurs through a combination of tails. In eukaryotes, this almost all known biological processes, including translational repression, deadenylation, decapping and process is catalysed by the cell growth, proliferation and differentiation, as well 5ʹ‑to‑3ʹ mRNA degradation5,6 (FIG. 1). The GW182 pro- consecutive but partially as organismal metabolism and development1. The teins play a central part in this process and are among redundant action of two 5,6 cytoplasmic deadenylase number of miRNAs encoded within the genomes of the most extensively studied AGO partners . -

Integrating Single-Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights Into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle
animals Article Integrating Single-Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle Masoumeh Naserkheil 1 , Abolfazl Bahrami 1 , Deukhwan Lee 2,* and Hossein Mehrban 3 1 Department of Animal Science, University College of Agriculture and Natural Resources, University of Tehran, Karaj 77871-31587, Iran; [email protected] (M.N.); [email protected] (A.B.) 2 Department of Animal Life and Environment Sciences, Hankyong National University, Jungang-ro 327, Anseong-si, Gyeonggi-do 17579, Korea 3 Department of Animal Science, Shahrekord University, Shahrekord 88186-34141, Iran; [email protected] * Correspondence: [email protected]; Tel.: +82-31-670-5091 Received: 25 August 2020; Accepted: 6 October 2020; Published: 9 October 2020 Simple Summary: Hanwoo is an indigenous cattle breed in Korea and popular for meat production owing to its rapid growth and high-quality meat. Its yearling weight and carcass traits (backfat thickness, carcass weight, eye muscle area, and marbling score) are economically important for the selection of young and proven bulls. In recent decades, the advent of high throughput genotyping technologies has made it possible to perform genome-wide association studies (GWAS) for the detection of genomic regions associated with traits of economic interest in different species. In this study, we conducted a weighted single-step genome-wide association study which combines all genotypes, phenotypes and pedigree data in one step (ssGBLUP). It allows for the use of all SNPs simultaneously along with all phenotypes from genotyped and ungenotyped animals. Our results revealed 33 relevant genomic regions related to the traits of interest. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Generation of H11-Albumin-Rtta Transgenic Mice: a Tool for Inducible Gene Expression in the Liver
INVESTIGATION Generation of H11-albumin-rtTA Transgenic Mice: A Tool for Inducible Gene Expression in the Liver Yu-Shan Li,*,1 Ran-Ran Meng,†,1 Xiu Chen,‡ Cui-Ling Shang,§ Hong-Bin Li,* Tao-Jun Zhang,† Hua-Yang Long,† Hui-Qi Li,† Yi-Jing Wang,† and Feng-Chao Wang†,2 *The School of Public Health, Xinxiang Medical University, Xinxiang, Henan, China 453003, †Transgenic Animal Center, National Institute of Biological Sciences, Beijing, China 102206, ‡Department of Pharmacy, Heze University, Heze, § Shandong, China, 274015, and Department of Reproductive Medicine, The Third Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan, China 453003 ABSTRACT The modification of the mouse genome by site-specific gene insertion of transgenes and other KEYWORDS genetic elements allows the study of gene function in different developmental stages and in the pathogenesis Hipp11 of diseases. Here, we generated a “genomic safe harbor” Hipp11 (H11)locus-specific knock-in transgenic albumin mouse line in which the albumin promoter is used to drive the expression of the reverse tetracycline promoter transactivator (rtTA) in the liver. The newly generated H11-albumin-rtTA transgenic mice were bred with doxycycline tetracycline-operator-Histone-2B-green fluorescent protein (TetO-H2BGFP) mice to assess inducibility and Tet-on system tissue-specificity. Expression of the H2BGFP fusion protein was observed exclusively upon doxycycline (Dox) induction in the liver of H11-albumin-rtTA/TetO-H2BGFP double transgenic mice. To further analyze the ability of the Dox-inducible H11-albumin-rtTA mice to implement conditional DNA recombination, H11- albumin-rtTA transgenic mice were crossed with TetO-Cre and Ai14 mice to generate H11-albumin-rtTA/ TetO-Cre/Ai14 triple transgenic mice. -

Open Data for Differential Network Analysis in Glioma
International Journal of Molecular Sciences Article Open Data for Differential Network Analysis in Glioma , Claire Jean-Quartier * y , Fleur Jeanquartier y and Andreas Holzinger Holzinger Group HCI-KDD, Institute for Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2/V, 8036 Graz, Austria; [email protected] (F.J.); [email protected] (A.H.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 27 October 2019; Accepted: 3 January 2020; Published: 15 January 2020 Abstract: The complexity of cancer diseases demands bioinformatic techniques and translational research based on big data and personalized medicine. Open data enables researchers to accelerate cancer studies, save resources and foster collaboration. Several tools and programming approaches are available for analyzing data, including annotation, clustering, comparison and extrapolation, merging, enrichment, functional association and statistics. We exploit openly available data via cancer gene expression analysis, we apply refinement as well as enrichment analysis via gene ontology and conclude with graph-based visualization of involved protein interaction networks as a basis for signaling. The different databases allowed for the construction of huge networks or specified ones consisting of high-confidence interactions only. Several genes associated to glioma were isolated via a network analysis from top hub nodes as well as from an outlier analysis. The latter approach highlights a mitogen-activated protein kinase next to a member of histondeacetylases and a protein phosphatase as genes uncommonly associated with glioma. Cluster analysis from top hub nodes lists several identified glioma-associated gene products to function within protein complexes, including epidermal growth factors as well as cell cycle proteins or RAS proto-oncogenes. -

The Mechanism of Eukaryotic Translation Initiation and Principles of Its Regulation
REVIEWS POST-TRANSCRIPTIONAL CONTROL The mechanism of eukaryotic translation initiation and principles of its regulation Richard J. Jackson*, Christopher U. T. Hellen‡ and Tatyana V. Pestova‡ Abstract | Protein synthesis is principally regulated at the initiation stage (rather than during elongation or termination), allowing rapid, reversible and spatial control of gene expression. Progress over recent years in determining the structures and activities of initiation factors, and in mapping their interactions in ribosomal initiation complexes, have advanced our understanding of the complex translation initiation process. These developments have provided a solid foundation for studying the regulation of translation initiation by mechanisms that include the modulation of initiation factor activity (which affects almost all scanning-dependent initiation) and through sequence-specific RNA-binding proteins and microRNAs (which affect individual mRNAs). Met Translation initiation is the process of assembly of significance is particularly high, and we include evi- Met-tRNA i The unique initiator tRNA, elongation-competent 80S ribosomes, in which the ini- dence from lower eukaryotes only when it enhances our aminoacylated with tiation codon is base-paired with the anticodon loop understanding of the mechanisms in vertebrates. methionine, that is used to Met 1 of initiator tRNA (Met-tRNA i) in the ribosomal P-site . initiate protein synthesis. Mechanism of 5′ end-dependent initiation Its anticodon is complementary It requires at least nine eukaryotic initiation factors to the AUG initiation codon; (eIFs; TABLE 1) and comprises two steps: the formation The canonical mechanism of translation initiation can be it forms a specific ternary of 48S initiation complexes with established codon– divided into several stages (FIG.1), as described below. -

Comparative Transcriptomics Reveals Similarities and Differences
Seifert et al. BMC Cancer (2015) 15:952 DOI 10.1186/s12885-015-1939-9 RESEARCH ARTICLE Open Access Comparative transcriptomics reveals similarities and differences between astrocytoma grades Michael Seifert1,2,5*, Martin Garbe1, Betty Friedrich1,3, Michel Mittelbronn4 and Barbara Klink5,6,7 Abstract Background: Astrocytomas are the most common primary brain tumors distinguished into four histological grades. Molecular analyses of individual astrocytoma grades have revealed detailed insights into genetic, transcriptomic and epigenetic alterations. This provides an excellent basis to identify similarities and differences between astrocytoma grades. Methods: We utilized public omics data of all four astrocytoma grades focusing on pilocytic astrocytomas (PA I), diffuse astrocytomas (AS II), anaplastic astrocytomas (AS III) and glioblastomas (GBM IV) to identify similarities and differences using well-established bioinformatics and systems biology approaches. We further validated the expression and localization of Ang2 involved in angiogenesis using immunohistochemistry. Results: Our analyses show similarities and differences between astrocytoma grades at the level of individual genes, signaling pathways and regulatory networks. We identified many differentially expressed genes that were either exclusively observed in a specific astrocytoma grade or commonly affected in specific subsets of astrocytoma grades in comparison to normal brain. Further, the number of differentially expressed genes generally increased with the astrocytoma grade with one major exception. The cytokine receptor pathway showed nearly the same number of differentially expressed genes in PA I and GBM IV and was further characterized by a significant overlap of commonly altered genes and an exclusive enrichment of overexpressed cancer genes in GBM IV. Additional analyses revealed a strong exclusive overexpression of CX3CL1 (fractalkine) and its receptor CX3CR1 in PA I possibly contributing to the absence of invasive growth. -
Download Validation Data
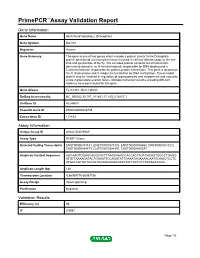
PrimePCR™Assay Validation Report Gene Information Gene Name dachshund homolog 2 (Drosophila) Gene Symbol DACH2 Organism Human Gene Summary This gene is one of two genes which encode a protein similar to the Drosophila protein dachshund a transcription factor involved in cell fate determination in the eye limb and genital disc of the fly. The encoded protein contains two characteristic dachshund domains: an N-terminal domain responsible for DNA binding and a C-terminal domain responsible for protein-protein interactions. This gene is located on the X chromosome and is subject to inactivation by DNA methylation. The encoded protein may be involved in regulation of organogenesis and myogenesis and may play a role in premature ovarian failure. Multiple transcript variants encoding different isoforms have been found for this gene. Gene Aliases FLJ31391, MGC138545 RefSeq Accession No. NC_000023.10, NT_011651.17, NG_012817.1 UniGene ID Hs.86603 Ensembl Gene ID ENSG00000126733 Entrez Gene ID 117154 Assay Information Unique Assay ID qHsaCID0009489 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000373131, ENST00000373125, ENST00000508860, ENST00000510272, ENST00000484479, ENST00000344497, ENST00000400297 Amplicon Context Sequence AGCAAGTGGAGCAGGCACTTAAGCAAGCCACCACTAGTGACAGTGGCCTGAGG ATGTTAAAAGATACTGGAATTCCAGATATTGAAATAGAAAACAATGGGACTCCTC ATGATAGTGCTGCTATGCAAGGAGGTAACTATTACTGTTTAGAAATGGC Amplicon Length (bp) 126 Chromosome Location X:86069775-86087136 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) -

Nº Ref Uniprot Proteína Péptidos Identificados Por MS/MS 1 P01024
Document downloaded from http://www.elsevier.es, day 26/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Nº Ref Uniprot Proteína Péptidos identificados 1 P01024 CO3_HUMAN Complement C3 OS=Homo sapiens GN=C3 PE=1 SV=2 por 162MS/MS 2 P02751 FINC_HUMAN Fibronectin OS=Homo sapiens GN=FN1 PE=1 SV=4 131 3 P01023 A2MG_HUMAN Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 128 4 P0C0L4 CO4A_HUMAN Complement C4-A OS=Homo sapiens GN=C4A PE=1 SV=1 95 5 P04275 VWF_HUMAN von Willebrand factor OS=Homo sapiens GN=VWF PE=1 SV=4 81 6 P02675 FIBB_HUMAN Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 78 7 P01031 CO5_HUMAN Complement C5 OS=Homo sapiens GN=C5 PE=1 SV=4 66 8 P02768 ALBU_HUMAN Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 66 9 P00450 CERU_HUMAN Ceruloplasmin OS=Homo sapiens GN=CP PE=1 SV=1 64 10 P02671 FIBA_HUMAN Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 58 11 P08603 CFAH_HUMAN Complement factor H OS=Homo sapiens GN=CFH PE=1 SV=4 56 12 P02787 TRFE_HUMAN Serotransferrin OS=Homo sapiens GN=TF PE=1 SV=3 54 13 P00747 PLMN_HUMAN Plasminogen OS=Homo sapiens GN=PLG PE=1 SV=2 48 14 P02679 FIBG_HUMAN Fibrinogen gamma chain OS=Homo sapiens GN=FGG PE=1 SV=3 47 15 P01871 IGHM_HUMAN Ig mu chain C region OS=Homo sapiens GN=IGHM PE=1 SV=3 41 16 P04003 C4BPA_HUMAN C4b-binding protein alpha chain OS=Homo sapiens GN=C4BPA PE=1 SV=2 37 17 Q9Y6R7 FCGBP_HUMAN IgGFc-binding protein OS=Homo sapiens GN=FCGBP PE=1 SV=3 30 18 O43866 CD5L_HUMAN CD5 antigen-like OS=Homo -

Loss of DACH2 Is a Candidate Early Event in Breast and Ovarian Carcinogenesis
Loss of DACH2 is a candidate early event in breast and ovarian carcinogenesis By Rania Chehade A thesis submitted in conformity with the requirements for Degree of Master of Science Medical Biophysics Department University of Toronto Copyright by Rania Chehade 2013 Loss of DACH2 is a candidate early event in breast and ovarian carcinogenesis Rania Chehade Master of Science Medical Biophysics Department University of Toronto 2013 Abstract Mechanistic insights into how enduring menstrual cycle hormonal signaling promotes tumorigenesis are emerging. We performed a genome-wide screen in primary epithelial cells to identify hormonally-regulated candidates that initiate pro-tumorigenic phenotypes in normal cells. One candidate, DACH2, has been described as part of a network that regulates organogenesis during development. In vitro, we find that DACH2 expression is regulated by estrogen and progesterone in a dosage dependent manner. Lentiviral-mediated shRNA silencing of DACH2 in hormonally responsive tissues promotes expansion of cells with progenitor characteristics. Within gene expression profiles of fallopian tubes, DACH2 is a member of a minimal gene classifier that distinguishes follicular versus luteal phases. Decreased DACH2 is characteristic of luteal-phase tubal cells and the majority of ovarian serous carcinomas. These studies suggest that DACH2 may orchestrate a physiological program that, when deregulated, locks cells in a progenitor-state, induces uncontrolled proliferation, and predisposes cells for breast and ovarian carcinogenesis. ii Acknowledgements Graduate school has been a bountiful journey of learning opportunities in academic, work and life relationships. I have developed various skills and I have matured both as a researcher and a human being. First and foremost, I would like to acknowledge my supervisor Dr.