DNA Replication During Acute MEK Inhibition Drives Acquisition of Resistance Through Amplification of the BRAF Oncogene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(AZD6244) in an in Vivo Model of Childhood Astrocytoma
Author Manuscript Published OnlineFirst on October 16, 2013; DOI: 10.1158/1078-0432.CCR-13-0842 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Development, Characterization, and Reversal of Acquired Resistance to the MEK1 Inhibitor Selumetinib (AZD6244) in an In Vivo Model of Childhood Astrocytoma Hemant K. Bid1, Aaron Kibler1, Doris A. Phelps1, Sagymbek Manap1, Linlin Xiao1, Jiayuh Lin1, David Capper2, Duane Oswald1, Brian Geier1, Mariko DeWire1,5, Paul D. Smith3, Raushan T. Kurmasheva1, Xiaokui Mo4, Soledad Fernandez4, and Peter J. Houghton1*. 1Center for Childhood Cancer & Blood Diseases, Nationwide Children’s Hospital, Columbus, OH 43205 2Institut of Pathology, Department Neuropathology, Ruprecht-Karls University and Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Astrazeneca Ltd., Oncology iMed, Macclesfield, U.K. 4Center for Biostatistics, The Ohio State University, Columbus, OH 43221 5 Present address: Cancer and Blood Diseases Institute, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229 Correspondence to Peter J. Houghton, Ph.D. Center for Childhood Cancer & Blood Diseases Nationwide Children’s Hospital 700 Children’s Drive Columbus, OH 43205 Ph: 614-355-2633 Fx: 614-355-2792 [email protected] Running head: Acquired resistance to MEK Inhibition in astrocytoma models. Conflict of Interest Statement: The authors consider that there is no actual or perceived conflict of interest. Dr. Paul D. Smith is an employee of Astrazeneca. 1 Downloaded from clincancerres.aacrjournals.org on September 30, 2021. © 2013 American Association for Cancer Research. Author Manuscript Published OnlineFirst on October 16, 2013; DOI: 10.1158/1078-0432.CCR-13-0842 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -

Could Hbx Protein Expression Affect Signal Pathway Inhibition by Gefitinib Or Selumetinib, a MEK Inhibitor, in Hepatocellular Carcinoma Cell Lines?
ORIGINAL ARTICLE Oncology & Hematology DOI: 10.3346/jkms.2011.26.2.214 • J Korean Med Sci 2011; 26: 214-221 Could HBx Protein Expression Affect Signal Pathway Inhibition by Gefitinib or Selumetinib, a MEK Inhibitor, in Hepatocellular Carcinoma Cell Lines? Yoon Kyung Park1, Kang Mo Kim1, Hepatitis B virus X (HBx) protein has been known to play an important role in development Young-Joo Lee2, Ki-Hun Kim2, of hepatocellular carcinoma (HCC). The aim of this study is to find out whether HBx Sung-Gyu Lee2, Danbi Lee1, protein expression affects antiproliferative effect of an epidermal growth factor receptor- Ju Hyun Shim1, Young-Suk Lim1, tyrosine kinase (EGFR-TK) inhibitor and a MEK inhibitor in HepG2 and Huh-7 cell lines. We 1 1 Han Chu Lee , Young-Hwa Chung , established HepG2 and Huh-7 cells transfected stably with HBx gene. HBx protein 1 1 Yung Sang Lee , and Dong Jin Suh expression increased pERK and pAkt expression as well as β-catenin activity in both cells. Departments of 1Internal Medicine and 2Surgery, Gefitinib (EGFR-TK inhibitor) inhibited pERK and pAkt expression andβ -catenin activity in Asan Medical Center, University of Ulsan College of both cells. Selumetinib (MEK inhibitor) reduced pERK level and β-catenin activity but pAkt Medicine, Seoul, Korea expression was rather elevated by selumetinib in these cells. Reduction of pERK levels was much stronger with selumetinib than gefitinib in both cells. The antiproliferative efficacy Received: 19 July 2010 Accepted: 2 November 2010 of selumetinib was more potent than that of gefitinib. However, the antiproliferative effect of gefitinib, as well as selumetinib, was not different between cell lines with or Address for Correspondence: without HBx expression. -

MET Or NRAS Amplification Is an Acquired Resistance Mechanism to the Third-Generation EGFR Inhibitor Naquotinib
www.nature.com/scientificreports OPEN MET or NRAS amplifcation is an acquired resistance mechanism to the third-generation EGFR inhibitor Received: 5 October 2017 Accepted: 16 January 2018 naquotinib Published: xx xx xxxx Kiichiro Ninomiya1, Kadoaki Ohashi1,2, Go Makimoto1, Shuta Tomida3, Hisao Higo1, Hiroe Kayatani1, Takashi Ninomiya1, Toshio Kubo4, Eiki Ichihara2, Katsuyuki Hotta5, Masahiro Tabata4, Yoshinobu Maeda1 & Katsuyuki Kiura2 As a third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI), osimeritnib is the standard treatment for patients with non-small cell lung cancer harboring the EGFR T790M mutation; however, acquired resistance inevitably develops. Therefore, a next-generation treatment strategy is warranted in the osimertinib era. We investigated the mechanism of resistance to a novel EGFR-TKI, naquotinib, with the goal of developing a novel treatment strategy. We established multiple naquotinib-resistant cell lines or osimertinib-resistant cells, two of which were derived from EGFR-TKI-naïve cells; the others were derived from geftinib- or afatinib-resistant cells harboring EGFR T790M. We comprehensively analyzed the RNA kinome sequence, but no universal gene alterations were detected in naquotinib-resistant cells. Neuroblastoma RAS viral oncogene homolog (NRAS) amplifcation was detected in naquotinib-resistant cells derived from geftinib-resistant cells. The combination therapy of MEK inhibitors and naquotinib exhibited a highly benefcial efect in resistant cells with NRAS amplifcation, but the combination of MEK inhibitors and osimertinib had limited efects on naquotinib-resistant cells. Moreover, the combination of MEK inhibitors and naquotinib inhibited the growth of osimertinib-resistant cells, while the combination of MEK inhibitors and osimertinib had little efect on osimertinib-resistant cells. -

Lenvatinib in Advanced, Radioactive Iodine– Refractory, Differentiated Thyroid Carcinoma Kay T
Published OnlineFirst October 20, 2015; DOI: 10.1158/1078-0432.CCR-15-0923 CCR Drug Updates Clinical Cancer Research Lenvatinib in Advanced, Radioactive Iodine– Refractory, Differentiated Thyroid Carcinoma Kay T. Yeung1 and Ezra E.W. Cohen2 Abstract Management options are limited for patients with radioactive therapy for these patients. Median PFS of 18.3 months in the iodine refractory, locally advanced, or metastatic differentiated lenvatinib group was significantly improved from 3.6 months in thyroid carcinoma. Prior to 2015, sorafenib, a multitargeted the placebo group, with an HR of 0.21 (95% confidence interval, tyrosine kinase inhibitor, was the only approved treatment and 0.4–0.31; P < 0.0001). ORR was also significantly increased in the was associated with a median progression-free survival (PFS) of lenvatinib arm (64.7%) compared with placebo (1.5%). In this 11 months and overall response rate (ORR) of 12% in a phase III article, we will review the molecular mechanisms of lenvatinib, trial. Lenvatinib, a multikinase inhibitor with high potency the data from preclinical studies to the recent phase III clinical against VEGFR and FGFR demonstrated encouraging results in trial, and the biomarkers being studied to further guide patient phase II trials. Recently, the pivotal SELECT trial provided the selection and predict treatment response. Clin Cancer Res; 21(24); basis for the FDA approval of lenvatinib as a second targeted 5420–6. Ó2015 AACR. Introduction PI3K–mTOR pathways (reviewed in ref. 3). The signals ultimately converge in the nucleus, influencing transcription of oncogenic Thyroid carcinoma is the most common endocrine malignan- proteins including, but not limited to, NF-kB), hypoxia-induced cy, with an estimated incidence rate of 13.5 per 100,000 people factor 1 alpha unit (HIF1a), TGFb, VEGF, and FGF. -

MEK1/2 Inhibitor Selumetinib (AZD6244) Inhibits Growth of Ovarian Clear Cell Carcinoma in a PEA-15–Dependent Manner in a Mouse Xenograft Model
Published OnlineFirst December 5, 2011; DOI: 10.1158/1535-7163.MCT-11-0400 Molecular Cancer Preclinical Development Therapeutics MEK1/2 Inhibitor Selumetinib (AZD6244) Inhibits Growth of Ovarian Clear Cell Carcinoma in a PEA-15–Dependent Manner in a Mouse Xenograft Model Chandra Bartholomeusz1,2, Tetsuro Oishi1,2,6, Hitomi Saso1,2, Ugur Akar1,2, Ping Liu3, Kimie Kondo1,2, Anna Kazansky1,2, Savitri Krishnamurthy4, Jangsoon Lee1,2, Francisco J. Esteva1,2, Junzo Kigawa6, and Naoto T. Ueno1,2,5 Abstract Clear cell carcinoma (CCC) of the ovary tends to show resistance to standard chemotherapy, which results in poor survival for patients with CCC. Developing a novel therapeutic strategy is imperative to improve patient prognosis. Epidermal growth factor receptor (EGFR) is frequently expressed in epithelial ovarian cancer. One of the major downstream targets of the EGFR signaling cascade is extracellular signal–related kinase (ERK). PEA-15, a 15-kDa phosphoprotein, can sequester ERK in the cytoplasm. MEK1/2 plays a central role in integrating mitogenic signals into the ERK pathway. We tested the hypothesis that inhibition of the EGFR–ERK pathway suppresses tumorigenicity in CCC, and we investigated the role of PEA-15 in ERK-targeted therapy in CCC. We screened a panel of 4 CCC cell lines (RMG-I, SMOV-2, OVTOKO, and KOC-7c) and observed that the EGFR tyrosine kinase inhibitor erlotinib inhibited cell proliferation of EGFR-overexpressing CCC cell lines through partial dependence on the MEK/ERK pathway. Furthermore, erlotinib-sensitive cell lines were also sensitive to the MEK inhibitor selumetinib (AZD6244), which is under clinical development. -

Combinatorial Efficacy of Entospletinib and Chemotherapy in Patient-Derived Xenograft Models of Infant Acute Lymphoblastic Leukemia
Acute Lymphoblastic Leukemia SUPPLEMENTARY APPENDIX Combinatorial efficacy of entospletinib and chemotherapy in patient-derived xenograft models of infant acute lymphoblastic leukemia Joseph P. Loftus, 1* Anella Yahiaoui, 2* Patrick A Brown, 3 Lisa M. Niswander, 1 Asen Bagashev, 1 Min Wang, 2 Allyson Shauf, 2 Stacey Tannheimer 2 and Sarah K. Tasian 1,4 1Division of Oncology and Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Philadelphia, PA; 2Gilead Sci - ences, Foster City, CA; 3Department of Pediatrics, Division of Pediatric Hematology/Oncology, Johns Hopkins University and Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD and 4Department of Pediatrics and Abramson Cancer Center, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA *JPL and AY contributed equally as co-first authors. ©2021 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2019.241729 Received: October 28, 2019. Accepted: May 8, 2020. Pre-published: May 15, 2020. Correspondence: SARAH K. TASIAN - [email protected] SUPPLEMENTAL DATA SUPPLEMENTAL METHODS Cell Titer Glo viability assays The human B-acute lymphoblastic leukemia (B-ALL) cell lines NALM-6 (non-KMT2A- rearranged) and HB11;19, KOPN-8, and HB11;19 (all KMT2A-rearranged) were obtained from the German Collection of Microorganisms and Cell Cultures GmbH (Deutsche Sammlung von Mikroorganismen und Zellkulturen; DSMZ), validated by short tandem repeat identity testing, and confirmed to be Mycoplasma-free. ALL cells were incubated in vitro with dimethyl sulfoxide or increasing concentrations of entospletinib (Gilead Sciences), fostamatinib (Selleckchem), dasatinib (LC Labs), selumetinib (LC Labs), and/or dexamethasone as indicated in Supplemental Figure 1 for 72 hours, then assessed for cell viability via Cell Titer Glo luminescent assays (Promega) according to manufacturer’s instructions using an IVIS Spectrum bioluminescent imaging instrument and Living Image software (PerkinElmer) (1, 2). -
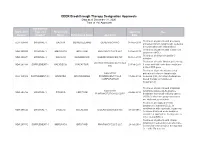
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

Trastuzumab Emtansine (T-DM1)
Published OnlineFirst September 11, 2018; DOI: 10.1158/1078-0432.CCR-18-1590 Research Article Clinical Cancer Research Trastuzumab Emtansine (T-DM1) in Patients with Previously Treated HER2-Overexpressing Metastatic Non–Small Cell Lung Cancer: Efficacy, Safety, and Biomarkers Solange Peters1, Rolf Stahel2, Lukas Bubendorf3, Philip Bonomi4, Augusto Villegas5, Dariusz M. Kowalski6, Christina S. Baik7, Dolores Isla8, Javier De Castro Carpeno9, Pilar Garrido10, Achim Rittmeyer11, Marcello Tiseo12, Christoph Meyenberg13, Sanne de Haas14, Lisa H. Lam15, Michael W. Lu15, and Thomas E. Stinchcombe16 Abstract Purpose: HER2-targeted therapy is not standard of care Results: Forty-nine patients received T-DM1 (29 IHC 2þ, for HER2-positive non–small cell lung cancer (NSCLC). This 20 IHC 3þ). No treatment responses were observed in the phase II study investigated efficacy and safety of the HER2- IHC 2þ cohort. Four partial responses were observed in the targeted antibody–drug conjugate trastuzumab emtansine IHC 3þ cohort (ORR, 20%; 95% confidence interval, 5.7%– (T-DM1) in patients with previously treated advanced 43.7%). Clinical benefit rates were 7% and 30% in the IHC HER2-overexpressing NSCLC. 2þ and 3þ cohorts, respectively. Response duration for the Patients and Methods: Eligible patients had HER2-over- responders was 2.9, 7.3, 8.3, and 10.8 months. Median expressing NSCLC (centrally tested IHC) and received progression-free survival and overall survival were similar previous platinum-based chemotherapy and targeted between cohorts. Three of 4 responders had HER2 gene therapy in the case of EGFR mutation or ALK gene amplification. No new safety signals were observed. rearrangement. Patients were divided into cohorts based Conclusions: T-DM1 showed a signal of activity in patients on HER2 IHC (2þ,3þ). -

Sorafenib/MEK Inhibitor Combination Inhibits Tumor Growth and the Wnt/Β‑Catenin Pathway in Xenograft Models of Hepatocellular Carcinoma
INTERNATIONAL JOURNAL OF ONCOLOGY 54: 1123-1133, 2019 Sorafenib/MEK inhibitor combination inhibits tumor growth and the Wnt/β‑catenin pathway in xenograft models of hepatocellular carcinoma HUNG HUYNH1, RICHARD ONG1, KAH YONG GOH1, LIEK YEOW LEE1, FLORIAN PUEHLER2, ARNE SCHOLZ2, OLIVER POLITZ2, DOMINIK MUMBERG2 and KARL ZIEGELBAUER2 1Laboratory of Molecular Endocrinology, Division of Molecular and Cellular Research, National Cancer Centre, Singapore 169610, Republic of Singapore; 2R&D Pharmaceuticals, Bayer AG, D‑13353 Berlin, Germany Received September 14, 2018; Accepted January 8, 2019 DOI: 10.3892/ijo.2019.4693 Abstract. Mutations affecting the Wnt/β-catenin pathway and refametinib inhibits the growth of naïve and sorafenib have been identified in 26‑40% of hepatocellular carcinoma resistant HCC tumors in association with active suppression of (HCC) cases. Aberrant activation of this pathway leads to β‑catenin signaling regardless of β‑catenin mutational status. uncontrolled cell proliferation and survival. Thus, identifying Thus, the sorafenib/MEK inhibitor combination may represent Wnt/β‑catenin pathway inhibitors may benefit a subset of an alternative treatment for patients with HCC whose tumors patients with HCC. In the present study, the effects of sorafenib develop resistance to sorafenib therapy. and a MEK inhibitor on tumor growth and Wnt/β-catenin signaling in HCC models were evaluated. A β‑catenin mutant Introduction and β‑catenin wild‑type HCC models were treated once daily with i) 10 mg/kg sorafenib, ii) 15 mg/kg refametinib Hepatocellular carcinoma (HCC) is the second most common (or 25 mg/kg selumetinib), or iii) sorafenib/refametinib. cause of cancer mortality worldwide, and accounts for Western blotting was employed to determine changes in 500,000 to 1 million deaths annually worldwide (1). -

Effect of Kinase Inhibitors on the Technetium-99M Uptake Into Thyroid Carcinoma Cells in Vitro ANNA ANSCHLAG 1, BRANDON H
in vivo 35 : 721-729 (2021) doi:10.21873/invivo.12313 Effect of Kinase Inhibitors on the Technetium-99m Uptake into Thyroid Carcinoma Cells In Vitro ANNA ANSCHLAG 1, BRANDON H. GREENE 2, LORIANNA KÖNNEKER 3, MARKUS LUSTER 4, JAMES NAGARAJAH 5, SABINE WÄCHTER 6, ANNETTE WUNDERLICH 6 and ANDREAS PFESTROFF 4 1Department of Child and Adolescent Psychiatry, University Hospital of Marburg, Marburg, Germany; 2Bio 21 Institute, CSL Limited Melbourne, Parkville, VIC, Australia; 3Department of Virology, Hospital Nordwest Frankfurt, Frankfurt, Germany; 4Department of Nuclear Medicine, University Hospital of Marburg, Marburg, Germany; 5Department of Nuclear Medicine, Klinikum Rechts der Isar, Technical University of Munich, Munich, Germany; 6Department of Visceral, Thoracic and Vascular Surgery, University Hospital of Marburg, Marburg, Germany Abstract. Background/Aim: We evaluated the potential of the distribution between the genders, and above 45,000 deaths kinase inhibitors sorafenib, lenvatinib and selumetinib on were recorded globally due to malignant thyroid disease (2). increasing the uptake of technetium-99m into thyroid cancer Its manifestations can be subdivided into different categories. cells. Materials and Methods: Four established cell lines and Differentiated thyroid carcinomas (DTC), accounting for about three patient’s cell cultures were treated with 0.1, 1 and 5μM 85% of all thyroid cancers, derive from the follicular of sorafenib, lenvatinib and selumetinib for 72 hours. After epithelium and include papillary (PTC) as well as follicular incubation with 1 MBq of technetium-99m, the radioactivity thyroid carcinoma (FTC), and a couple of rare subtypes. uptake was measured. Results: The experiments showed Medullary thyroid cancers (MTC) are developed from heterogeneous results. Maximum technetium-99m uptake calcitonin producing C-cells, whereas the highly aggressive increases of 312% (sorafenib), 326% (lenvatinib) and 759% anaplastic thyroid carcinomas (ATC) are composed of (selumetinib) were obtained using the highest applied dedifferentiated tissue. -

A Perspective from Clinical Trials
biomolecules Review Development of Possible Next Line of Systemic Therapies for Gemcitabine-Resistant Biliary Tract Cancers: A Perspective from Clinical Trials Nai-Jung Chiang 1,2, Li-Tzong Chen 1,2,3, Yan-Shen Shan 4,5, Chun-Nan Yeh 6,* and Ming-Huang Chen 7,8,* 1 National Institute of Cancer Research, National Health Research Institutes, Tainan 704, Taiwan; [email protected] (N.-J.C.); [email protected] (L.-T.C.) 2 Department of Oncology, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan 704, Taiwan 3 Department of Internal Medicine, Kaohsiung Medical University Hospital and Kaohsiung Medical University, Kaohsiung 807, Taiwan 4 Institute of Clinical Medicine, College of Medicine, National Cheng Kung University, Tainan 704, Taiwan; [email protected] 5 Department of Surgery, National Cheng Kung University Hospital, Tainan 704, Taiwan 6 Department of General Surgery and Liver Research Center, Chang Gung Memorial Hospital, Linkou Branch, Chang Gung University, Taoyuan 333, Taiwan 7 Center for Immuno-Oncology, Department of Oncology, Taipei Veterans General Hospital, Taipei 112, Taiwan 8 School of Medicine, National Yang Ming University, Taipei 112, Taiwan * Correspondence: [email protected] (C.-N.Y.); [email protected] (M.-H.C.); Tel.: +886-33281200 (C.-N.Y.); +886-28712121 (ext. 2508) (M.-H.C.); Fax: +886-33285818 (C.-N.Y.); +886-28732131 (M.-H.C.) Abstract: Biliary tract cancer (BTC) compromises a heterogenous group of tumors with poor prog- noses. Curative surgery remains the first choice for localized disease; however, most BTC pa- tients have had unresectable or metastatic disease. -

Phase Ib Results of the Rational Combination Of
Published OnlineFirst July 24, 2018; DOI: 10.1158/0008-5472.CAN-18-0316 Cancer Translational Science Research Phase Ib Results of the Rational Combination of Selumetinib and Cyclosporin A in Advanced Solid Tumors with an Expansion Cohort in Metastatic Colorectal Cancer Anuradha Krishnamurthy1, Arvind Dasari2, Anne M. Noonan3, Janice M. Mehnert4, Albert C. Lockhart5, Stephen Leong1, Anna Capasso1, Mark N. Stein4, Hanna K. Sanoff6, James J. Lee7, Aaron Hansen8, Usha Malhotra4, Sarah Rippke1, Daniel L. Gustafson9, Todd M. Pitts1, Kim Ellison1, S. Lindsey Davis1, Wells A. Messersmith1, S. Gail Eckhardt1,10, and Christopher H. Lieu1 Abstract MEK inhibition is of interest in cancer drug development, sion phases, respectively. The most common adverse events but clinical activity in metastatic colorectal cancer (mCRC) has and grade 3/4 toxicities were rash, hypertension, and edema. been limited. Preclinical studies demonstrated Wnt pathway Three dose-limiting toxicities (grade 3 hypertension, rash, overexpression in KRAS-mutant cell lines resistant to the MEK and increased creatinine) were reported. The MTD was inhibitor, selumetinib. The combination of selumetinib selumetinib 75 mg twice daily and cyclosporin A 2 mg/kg and cyclosporin A, a noncanonical Wnt pathway modulator, twice daily on a 28-day cycle. KRAS stratification did not demonstrated antitumor activity in mCRC patient-derived identify any differences in response between KRAS-WT xenografts. To translate these results, we conducted a NCI and KRAS-MT cancers. Two partial responses, 18 stable Cancer Therapy Evaluation Program–approved multicenter disease, and 10 progressive disease responses were observed. phase I/IB trial (NCT02188264) of the combination Combination selumetinib and cyclosporin A is well toler- of selumetinib and cyclosporin A.