Combinatorial Efficacy of Entospletinib and Chemotherapy in Patient-Derived Xenograft Models of Infant Acute Lymphoblastic Leukemia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(AZD6244) in an in Vivo Model of Childhood Astrocytoma
Author Manuscript Published OnlineFirst on October 16, 2013; DOI: 10.1158/1078-0432.CCR-13-0842 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Development, Characterization, and Reversal of Acquired Resistance to the MEK1 Inhibitor Selumetinib (AZD6244) in an In Vivo Model of Childhood Astrocytoma Hemant K. Bid1, Aaron Kibler1, Doris A. Phelps1, Sagymbek Manap1, Linlin Xiao1, Jiayuh Lin1, David Capper2, Duane Oswald1, Brian Geier1, Mariko DeWire1,5, Paul D. Smith3, Raushan T. Kurmasheva1, Xiaokui Mo4, Soledad Fernandez4, and Peter J. Houghton1*. 1Center for Childhood Cancer & Blood Diseases, Nationwide Children’s Hospital, Columbus, OH 43205 2Institut of Pathology, Department Neuropathology, Ruprecht-Karls University and Clinical Cooperation Unit Neuropathology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Astrazeneca Ltd., Oncology iMed, Macclesfield, U.K. 4Center for Biostatistics, The Ohio State University, Columbus, OH 43221 5 Present address: Cancer and Blood Diseases Institute, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229 Correspondence to Peter J. Houghton, Ph.D. Center for Childhood Cancer & Blood Diseases Nationwide Children’s Hospital 700 Children’s Drive Columbus, OH 43205 Ph: 614-355-2633 Fx: 614-355-2792 [email protected] Running head: Acquired resistance to MEK Inhibition in astrocytoma models. Conflict of Interest Statement: The authors consider that there is no actual or perceived conflict of interest. Dr. Paul D. Smith is an employee of Astrazeneca. 1 Downloaded from clincancerres.aacrjournals.org on September 30, 2021. © 2013 American Association for Cancer Research. Author Manuscript Published OnlineFirst on October 16, 2013; DOI: 10.1158/1078-0432.CCR-13-0842 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -

Could Hbx Protein Expression Affect Signal Pathway Inhibition by Gefitinib Or Selumetinib, a MEK Inhibitor, in Hepatocellular Carcinoma Cell Lines?
ORIGINAL ARTICLE Oncology & Hematology DOI: 10.3346/jkms.2011.26.2.214 • J Korean Med Sci 2011; 26: 214-221 Could HBx Protein Expression Affect Signal Pathway Inhibition by Gefitinib or Selumetinib, a MEK Inhibitor, in Hepatocellular Carcinoma Cell Lines? Yoon Kyung Park1, Kang Mo Kim1, Hepatitis B virus X (HBx) protein has been known to play an important role in development Young-Joo Lee2, Ki-Hun Kim2, of hepatocellular carcinoma (HCC). The aim of this study is to find out whether HBx Sung-Gyu Lee2, Danbi Lee1, protein expression affects antiproliferative effect of an epidermal growth factor receptor- Ju Hyun Shim1, Young-Suk Lim1, tyrosine kinase (EGFR-TK) inhibitor and a MEK inhibitor in HepG2 and Huh-7 cell lines. We 1 1 Han Chu Lee , Young-Hwa Chung , established HepG2 and Huh-7 cells transfected stably with HBx gene. HBx protein 1 1 Yung Sang Lee , and Dong Jin Suh expression increased pERK and pAkt expression as well as β-catenin activity in both cells. Departments of 1Internal Medicine and 2Surgery, Gefitinib (EGFR-TK inhibitor) inhibited pERK and pAkt expression andβ -catenin activity in Asan Medical Center, University of Ulsan College of both cells. Selumetinib (MEK inhibitor) reduced pERK level and β-catenin activity but pAkt Medicine, Seoul, Korea expression was rather elevated by selumetinib in these cells. Reduction of pERK levels was much stronger with selumetinib than gefitinib in both cells. The antiproliferative efficacy Received: 19 July 2010 Accepted: 2 November 2010 of selumetinib was more potent than that of gefitinib. However, the antiproliferative effect of gefitinib, as well as selumetinib, was not different between cell lines with or Address for Correspondence: without HBx expression. -

Management of Chronic Myelogenous Leukemia in Pregnancy
ANTICANCER RESEARCH 35: 1-12 (2015) Review Management of Chronic Myelogenous Leukemia in Pregnancy AMIT BHANDARI, KATRINA ROLEN and BINAY KUMAR SHAH Cancer Center and Blood Institute, St. Joseph Regional Medical Center, Lewiston, ID, U.S.A. Abstract. Discovery of tyrosine kinase inhibitors has led to Leukemia in pregnancy is a rare condition, with an annual improvement in survival of chronic myelogenous leukemia incidence of 1-2/100,000 pregnancies (8). Since the first (CML) patients. Many young CML patients encounter administration of imatinib (the first of the TKIs) to patients pregnancy during their lifetime. Tyrosine kinase inhibitors with CML in June 1998, it is estimated that there have now inhibit several proteins that are known to have important been 250,000 patient years of exposure to the drug (mostly functions in gonadal development, implantation and fetal in patients with CML) (9). TKIs not only target BCR-ABL development, thus increasing the risk of embryo toxicities. tyrosine kinase but also c-kit, platelet derived growth factors Studies have shown imatinib to be embryotoxic in animals with receptors α and β (PDGFR-α/β), ARG and c-FMS (10). varying effects in fertility. Since pregnancy is rare in CML, Several of these proteins are known to have functions that there are no randomized controlled trials to address the may be important in gonadal development, implantation and optimal management of this condition. However, there are fetal development (11-15). Despite this fact, there is still only several case reports and case series on CML in pregnancy. At limited information on the effects of imatinib on fertility the present time, there is no consensus on how to manage and/or pregnancy. -

MET Or NRAS Amplification Is an Acquired Resistance Mechanism to the Third-Generation EGFR Inhibitor Naquotinib
www.nature.com/scientificreports OPEN MET or NRAS amplifcation is an acquired resistance mechanism to the third-generation EGFR inhibitor Received: 5 October 2017 Accepted: 16 January 2018 naquotinib Published: xx xx xxxx Kiichiro Ninomiya1, Kadoaki Ohashi1,2, Go Makimoto1, Shuta Tomida3, Hisao Higo1, Hiroe Kayatani1, Takashi Ninomiya1, Toshio Kubo4, Eiki Ichihara2, Katsuyuki Hotta5, Masahiro Tabata4, Yoshinobu Maeda1 & Katsuyuki Kiura2 As a third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI), osimeritnib is the standard treatment for patients with non-small cell lung cancer harboring the EGFR T790M mutation; however, acquired resistance inevitably develops. Therefore, a next-generation treatment strategy is warranted in the osimertinib era. We investigated the mechanism of resistance to a novel EGFR-TKI, naquotinib, with the goal of developing a novel treatment strategy. We established multiple naquotinib-resistant cell lines or osimertinib-resistant cells, two of which were derived from EGFR-TKI-naïve cells; the others were derived from geftinib- or afatinib-resistant cells harboring EGFR T790M. We comprehensively analyzed the RNA kinome sequence, but no universal gene alterations were detected in naquotinib-resistant cells. Neuroblastoma RAS viral oncogene homolog (NRAS) amplifcation was detected in naquotinib-resistant cells derived from geftinib-resistant cells. The combination therapy of MEK inhibitors and naquotinib exhibited a highly benefcial efect in resistant cells with NRAS amplifcation, but the combination of MEK inhibitors and osimertinib had limited efects on naquotinib-resistant cells. Moreover, the combination of MEK inhibitors and naquotinib inhibited the growth of osimertinib-resistant cells, while the combination of MEK inhibitors and osimertinib had little efect on osimertinib-resistant cells. -

DNA Replication During Acute MEK Inhibition Drives Acquisition of Resistance Through Amplification of the BRAF Oncogene
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.23.436572; this version posted March 23, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Acquisition of MEKi resistance during DNA replication in drug Channathodiyil et al. DNA replication during acute MEK inhibition drives acquisition of resistance through amplification of the BRAF oncogene Prasanna Channathodiyil1,2, Anne Segonds-Pichon3, Paul D. Smith4, Simon J. Cook5 and Jonathan Houseley1,6,* 1 Epigenetics Programme, Babraham Institute, Cambridge, UK 2 ORCID: 0000-0002-9381-6089 3 Babraham Bioinformatics, Babraham Institute, Cambridge, UK. ORCID: 0000-0002-8369- 4882 4 Oncology R&D, AstraZeneca CRUK Cambridge Institute, Cambridge, UK 5 Signalling programme, Babraham Institute, Cambridge, UK. ORCID: 0000-0001-9087-1616 6 ORCID: 0000-0001-8509-1500 * Corresponding author: [email protected] Abstract Mutations and gene amplifications that confer drug resistance emerge frequently during chemotherapy, but their mechanism and timing is poorly understood. Here, we investigate BRAFV600E amplification events that underlie resistance to the MEK inhibitor selumetinib (AZD6244/ARRY-142886) in COLO205 cells. We find that de novo focal BRAF amplification is the primary path to resistance irrespective of pre-existing amplifications. Although selumetinib causes long-term G1 arrest, we observe that cells stochastically re-enter the cell cycle during treatment without reactivation of ERK1/2 or induction of a normal proliferative gene expression programme. Genes encoding DNA replication and repair factors are downregulated during G1 arrest, but many are transiently induced when cells escape arrest and enter S and G2. -

Lenvatinib in Advanced, Radioactive Iodine– Refractory, Differentiated Thyroid Carcinoma Kay T
Published OnlineFirst October 20, 2015; DOI: 10.1158/1078-0432.CCR-15-0923 CCR Drug Updates Clinical Cancer Research Lenvatinib in Advanced, Radioactive Iodine– Refractory, Differentiated Thyroid Carcinoma Kay T. Yeung1 and Ezra E.W. Cohen2 Abstract Management options are limited for patients with radioactive therapy for these patients. Median PFS of 18.3 months in the iodine refractory, locally advanced, or metastatic differentiated lenvatinib group was significantly improved from 3.6 months in thyroid carcinoma. Prior to 2015, sorafenib, a multitargeted the placebo group, with an HR of 0.21 (95% confidence interval, tyrosine kinase inhibitor, was the only approved treatment and 0.4–0.31; P < 0.0001). ORR was also significantly increased in the was associated with a median progression-free survival (PFS) of lenvatinib arm (64.7%) compared with placebo (1.5%). In this 11 months and overall response rate (ORR) of 12% in a phase III article, we will review the molecular mechanisms of lenvatinib, trial. Lenvatinib, a multikinase inhibitor with high potency the data from preclinical studies to the recent phase III clinical against VEGFR and FGFR demonstrated encouraging results in trial, and the biomarkers being studied to further guide patient phase II trials. Recently, the pivotal SELECT trial provided the selection and predict treatment response. Clin Cancer Res; 21(24); basis for the FDA approval of lenvatinib as a second targeted 5420–6. Ó2015 AACR. Introduction PI3K–mTOR pathways (reviewed in ref. 3). The signals ultimately converge in the nucleus, influencing transcription of oncogenic Thyroid carcinoma is the most common endocrine malignan- proteins including, but not limited to, NF-kB), hypoxia-induced cy, with an estimated incidence rate of 13.5 per 100,000 people factor 1 alpha unit (HIF1a), TGFb, VEGF, and FGF. -

The Effects of Combination Treatments on Drug Resistance in Chronic Myeloid Leukaemia: an Evaluation of the Tyrosine Kinase Inhibitors Axitinib and Asciminib H
Lindström and Friedman BMC Cancer (2020) 20:397 https://doi.org/10.1186/s12885-020-06782-9 RESEARCH ARTICLE Open Access The effects of combination treatments on drug resistance in chronic myeloid leukaemia: an evaluation of the tyrosine kinase inhibitors axitinib and asciminib H. Jonathan G. Lindström and Ran Friedman* Abstract Background: Chronic myeloid leukaemia is in principle a treatable malignancy but drug resistance is lowering survival. Recent drug discoveries have opened up new options for drug combinations, which is a concept used in other areas for preventing drug resistance. Two of these are (I) Axitinib, which inhibits the T315I mutation of BCR-ABL1, a main source of drug resistance, and (II) Asciminib, which has been developed as an allosteric BCR-ABL1 inhibitor, targeting an entirely different binding site, and as such does not compete for binding with other drugs. These drugs offer new treatment options. Methods: We measured the proliferation of KCL-22 cells exposed to imatinib–dasatinib, imatinib–asciminib and dasatinib–asciminib combinations and calculated combination index graphs for each case. Moreover, using the median–effect equation we calculated how much axitinib can reduce the growth advantage of T315I mutant clones in combination with available drugs. In addition, we calculated how much the total drug burden could be reduced by combinations using asciminib and other drugs, and evaluated which mutations such combinations might be sensitive to. Results: Asciminib had synergistic interactions with imatinib or dasatinib in KCL-22 cells at high degrees of inhibition. Interestingly, some antagonism between asciminib and the other drugs was present at lower degrees on inhibition. -

MEK1/2 Inhibitor Selumetinib (AZD6244) Inhibits Growth of Ovarian Clear Cell Carcinoma in a PEA-15–Dependent Manner in a Mouse Xenograft Model
Published OnlineFirst December 5, 2011; DOI: 10.1158/1535-7163.MCT-11-0400 Molecular Cancer Preclinical Development Therapeutics MEK1/2 Inhibitor Selumetinib (AZD6244) Inhibits Growth of Ovarian Clear Cell Carcinoma in a PEA-15–Dependent Manner in a Mouse Xenograft Model Chandra Bartholomeusz1,2, Tetsuro Oishi1,2,6, Hitomi Saso1,2, Ugur Akar1,2, Ping Liu3, Kimie Kondo1,2, Anna Kazansky1,2, Savitri Krishnamurthy4, Jangsoon Lee1,2, Francisco J. Esteva1,2, Junzo Kigawa6, and Naoto T. Ueno1,2,5 Abstract Clear cell carcinoma (CCC) of the ovary tends to show resistance to standard chemotherapy, which results in poor survival for patients with CCC. Developing a novel therapeutic strategy is imperative to improve patient prognosis. Epidermal growth factor receptor (EGFR) is frequently expressed in epithelial ovarian cancer. One of the major downstream targets of the EGFR signaling cascade is extracellular signal–related kinase (ERK). PEA-15, a 15-kDa phosphoprotein, can sequester ERK in the cytoplasm. MEK1/2 plays a central role in integrating mitogenic signals into the ERK pathway. We tested the hypothesis that inhibition of the EGFR–ERK pathway suppresses tumorigenicity in CCC, and we investigated the role of PEA-15 in ERK-targeted therapy in CCC. We screened a panel of 4 CCC cell lines (RMG-I, SMOV-2, OVTOKO, and KOC-7c) and observed that the EGFR tyrosine kinase inhibitor erlotinib inhibited cell proliferation of EGFR-overexpressing CCC cell lines through partial dependence on the MEK/ERK pathway. Furthermore, erlotinib-sensitive cell lines were also sensitive to the MEK inhibitor selumetinib (AZD6244), which is under clinical development. -

Antibody–Drug Conjugates: the Last Decade
pharmaceuticals Review Antibody–Drug Conjugates: The Last Decade Nicolas Joubert 1,* , Alain Beck 2 , Charles Dumontet 3,4 and Caroline Denevault-Sabourin 1 1 GICC EA7501, Equipe IMT, Université de Tours, UFR des Sciences Pharmaceutiques, 31 Avenue Monge, 37200 Tours, France; [email protected] 2 Institut de Recherche Pierre Fabre, Centre d’Immunologie Pierre Fabre, 5 Avenue Napoléon III, 74160 Saint Julien en Genevois, France; [email protected] 3 Cancer Research Center of Lyon (CRCL), INSERM, 1052/CNRS 5286/UCBL, 69000 Lyon, France; [email protected] 4 Hospices Civils de Lyon, 69000 Lyon, France * Correspondence: [email protected] Received: 17 August 2020; Accepted: 10 September 2020; Published: 14 September 2020 Abstract: An armed antibody (antibody–drug conjugate or ADC) is a vectorized chemotherapy, which results from the grafting of a cytotoxic agent onto a monoclonal antibody via a judiciously constructed spacer arm. ADCs have made considerable progress in 10 years. While in 2009 only gemtuzumab ozogamicin (Mylotarg®) was used clinically, in 2020, 9 Food and Drug Administration (FDA)-approved ADCs are available, and more than 80 others are in active clinical studies. This review will focus on FDA-approved and late-stage ADCs, their limitations including their toxicity and associated resistance mechanisms, as well as new emerging strategies to address these issues and attempt to widen their therapeutic window. Finally, we will discuss their combination with conventional chemotherapy or checkpoint inhibitors, and their design for applications beyond oncology, to make ADCs the magic bullet that Paul Ehrlich dreamed of. Keywords: antibody–drug conjugate; ADC; bioconjugation; linker; payload; cancer; resistance; combination therapies 1. -
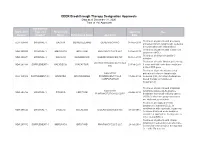
CDER Breakthrough Therapy Designation Approvals Data As of December 31, 2020 Total of 190 Approvals
CDER Breakthrough Therapy Designation Approvals Data as of December 31, 2020 Total of 190 Approvals Submission Application Type and Proprietary Approval Use Number Number Name Established Name Applicant Date Treatment of patients with previously BLA 125486 ORIGINAL-1 GAZYVA OBINUTUZUMAB GENENTECH INC 01-Nov-2013 untreated chronic lymphocytic leukemia in combination with chlorambucil Treatment of patients with mantle cell NDA 205552 ORIGINAL-1 IMBRUVICA IBRUTINIB PHARMACYCLICS LLC 13-Nov-2013 lymphoma (MCL) Treatment of chronic hepatitis C NDA 204671 ORIGINAL-1 SOVALDI SOFOSBUVIR GILEAD SCIENCES INC 06-Dec-2013 infection Treatment of cystic fibrosis patients age VERTEX PHARMACEUTICALS NDA 203188 SUPPLEMENT-4 KALYDECO IVACAFTOR 21-Feb-2014 6 years and older who have mutations INC in the CFTR gene Treatment of previously untreated NOVARTIS patients with chronic lymphocytic BLA 125326 SUPPLEMENT-60 ARZERRA OFATUMUMAB PHARMACEUTICALS 17-Apr-2014 leukemia (CLL) for whom fludarabine- CORPORATION based therapy is considered inappropriate Treatment of patients with anaplastic NOVARTIS lymphoma kinase (ALK)-positive NDA 205755 ORIGINAL-1 ZYKADIA CERITINIB 29-Apr-2014 PHARMACEUTICALS CORP metastatic non-small cell lung cancer (NSCLC) who have progressed on or are intolerant to crizotinib Treatment of relapsed chronic lymphocytic leukemia (CLL), in combination with rituximab, in patients NDA 206545 ORIGINAL-1 ZYDELIG IDELALISIB GILEAD SCIENCES INC 23-Jul-2014 for whom rituximab alone would be considered appropriate therapy due to other co-morbidities -

2021 Formulary List of Covered Prescription Drugs
2021 Formulary List of covered prescription drugs This drug list applies to all Individual HMO products and the following Small Group HMO products: Sharp Platinum 90 Performance HMO, Sharp Platinum 90 Performance HMO AI-AN, Sharp Platinum 90 Premier HMO, Sharp Platinum 90 Premier HMO AI-AN, Sharp Gold 80 Performance HMO, Sharp Gold 80 Performance HMO AI-AN, Sharp Gold 80 Premier HMO, Sharp Gold 80 Premier HMO AI-AN, Sharp Silver 70 Performance HMO, Sharp Silver 70 Performance HMO AI-AN, Sharp Silver 70 Premier HMO, Sharp Silver 70 Premier HMO AI-AN, Sharp Silver 73 Performance HMO, Sharp Silver 73 Premier HMO, Sharp Silver 87 Performance HMO, Sharp Silver 87 Premier HMO, Sharp Silver 94 Performance HMO, Sharp Silver 94 Premier HMO, Sharp Bronze 60 Performance HMO, Sharp Bronze 60 Performance HMO AI-AN, Sharp Bronze 60 Premier HDHP HMO, Sharp Bronze 60 Premier HDHP HMO AI-AN, Sharp Minimum Coverage Performance HMO, Sharp $0 Cost Share Performance HMO AI-AN, Sharp $0 Cost Share Premier HMO AI-AN, Sharp Silver 70 Off Exchange Performance HMO, Sharp Silver 70 Off Exchange Premier HMO, Sharp Performance Platinum 90 HMO 0/15 + Child Dental, Sharp Premier Platinum 90 HMO 0/20 + Child Dental, Sharp Performance Gold 80 HMO 350 /25 + Child Dental, Sharp Premier Gold 80 HMO 250/35 + Child Dental, Sharp Performance Silver 70 HMO 2250/50 + Child Dental, Sharp Premier Silver 70 HMO 2250/55 + Child Dental, Sharp Premier Silver 70 HDHP HMO 2500/20% + Child Dental, Sharp Performance Bronze 60 HMO 6300/65 + Child Dental, Sharp Premier Bronze 60 HDHP HMO -

Trastuzumab Emtansine (T-DM1)
Published OnlineFirst September 11, 2018; DOI: 10.1158/1078-0432.CCR-18-1590 Research Article Clinical Cancer Research Trastuzumab Emtansine (T-DM1) in Patients with Previously Treated HER2-Overexpressing Metastatic Non–Small Cell Lung Cancer: Efficacy, Safety, and Biomarkers Solange Peters1, Rolf Stahel2, Lukas Bubendorf3, Philip Bonomi4, Augusto Villegas5, Dariusz M. Kowalski6, Christina S. Baik7, Dolores Isla8, Javier De Castro Carpeno9, Pilar Garrido10, Achim Rittmeyer11, Marcello Tiseo12, Christoph Meyenberg13, Sanne de Haas14, Lisa H. Lam15, Michael W. Lu15, and Thomas E. Stinchcombe16 Abstract Purpose: HER2-targeted therapy is not standard of care Results: Forty-nine patients received T-DM1 (29 IHC 2þ, for HER2-positive non–small cell lung cancer (NSCLC). This 20 IHC 3þ). No treatment responses were observed in the phase II study investigated efficacy and safety of the HER2- IHC 2þ cohort. Four partial responses were observed in the targeted antibody–drug conjugate trastuzumab emtansine IHC 3þ cohort (ORR, 20%; 95% confidence interval, 5.7%– (T-DM1) in patients with previously treated advanced 43.7%). Clinical benefit rates were 7% and 30% in the IHC HER2-overexpressing NSCLC. 2þ and 3þ cohorts, respectively. Response duration for the Patients and Methods: Eligible patients had HER2-over- responders was 2.9, 7.3, 8.3, and 10.8 months. Median expressing NSCLC (centrally tested IHC) and received progression-free survival and overall survival were similar previous platinum-based chemotherapy and targeted between cohorts. Three of 4 responders had HER2 gene therapy in the case of EGFR mutation or ALK gene amplification. No new safety signals were observed. rearrangement. Patients were divided into cohorts based Conclusions: T-DM1 showed a signal of activity in patients on HER2 IHC (2þ,3þ).