Molecular and Evolutionary Aspects of the Protochordate Digestive System
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diverticular Disease 1
WGO Practice Guidelines Diverticular disease 1 World Gastroenterology Organisation Practice Guidelines: Diverticular Disease Core review team: Dr. T. Murphy Prof. R.H. Hunt Prof. M. Fried Dr. J.H. Krabshuis Contents 1 Definitions 2 Epidemiology 3 Etiology 4 Pathophysiology 5 Medical and surgical management 6 Other forms of diverticular disease 7 Global aspects 8 References 9 Useful web sites 10 WGO Practice Guidelines Committee members who contributed to this guideline 11 Queries and feedback 1 Definitions Diverticulum: • A sac-like protrusion of mucosa through the muscular colonic wall [1]. • Protrusion occurs in weak areas of the bowel wall through which blood vessels can penetrate. • Typically 5–10 mm in size. • Diverticula are really pseudodiverticular (false diverticula), as they contain only mucosa and submucosa covered by serosa. Diverticular disease consists of: • Diverticulosis: the presence of diverticula within the colon • Diverticulitis: inflammation of a diverticulum • Diverticular bleeding Types of diverticular disease: © World Gastroenterology Organisation, 2007 WGO Practice Guidelines Diverticular disease 2 • Simple (75%), with no complications • Complicated (25%), with abscesses, fistula, obstruction, peritonitis, and sepsis 2 Epidemiology Prevalence by age [1]: • Age 40: 5% • Age 60: 30% • Age 80: 65% Prevalence by sex: • Age < 50: more common in males • Age 50–70: slight preponderance in women • Age > 70: more common in women Diverticular disease in the young (< 40) Diverticular disease is far more frequent in older people, with only 2–5% of cases occurring in those under 40 years of age. In this younger age group, diverticular disease occurs more frequently in males, with obesity being a major risk factor (present in 84–96% of cases) [2,3]. -

Jejunoileal Diverticulitis: Big Trouble in Small Bowel
Jejunoileal Diverticulitis: Big Trouble in Small Bowel MICHAEL CARUSO DO, HAO LO MD EMERGENCY RADIOLOGY, UMASS MEMORIAL MEDICAL CENTER, WORCESTER, MA LEARNING OBJECTIVES: After excluding Meckel’s diverticulum, less than 30% of reported diverticula occurred in the CASE 1 1. Be aware of the relatively rare diagnoses of jejunoileal jejunum and ileum. HISTORY: 52 year old female with left upper quadrant pain. diverticulosis (non-Meckelian) and diverticulitis. Duodenal diverticula are approximately five times more common than jejunoileal diverticula. FINDINGS: 2.2 cm diverticulum extending from the distal ileum with adjacent induration of the mesenteric fat, consistent with an ileal diverticulitis. 2. Learn the epidemiology, pathophysiology, imaging findings and differential diagnosis of jejunoileal diverticulitis. Diverticulaare sac-like protrusions of the bowel wall, which may be composed of mucosa and submucosa only (pseudodiverticula) or of all CT Findings (2) layers of the bowel wall (true . Usually presents as a focal area of bowel wall thickening most prominent on the mesenteric side of AXIAL AXIAL CORONAL diverticula) the bowel with adjacent inflammation and/or abscess formation . Diverticulitis is the result of When abscess is present, CT findings may include relatively smooth margins, areas of low CASE 2 obstruction of the neck of the attenuation within the mass, rim enhancement after IV contrast administration, gas within the HISTORY: 62 year old female with epigastric and left upper quadrant pain. diverticulum, with subsequent mass, displacement of the surrounding structures, and edema of thickening of the surrounding fat inflammation, perforation and or fascial planes FINDINGS: 4.7 cm diameter out pouching seen arising from the proximal small bowel and associated with adjacent inflammatory stranding. -

Prey Capture and Digestion in the Carnivorous Sponge Asbestopluma Hypogea (Porifera: Demospongiae)
Zoomorphology (2004) 123:179–190 DOI 10.1007/s00435-004-0100-0 ORIGINAL ARTICLE Jean Vacelet · Eric Duport Prey capture and digestion in the carnivorous sponge Asbestopluma hypogea (Porifera: Demospongiae) Received: 2 May 2003 / Accepted: 17 March 2004 / Published online: 27 April 2004 Springer-Verlag 2004 Abstract Asbestopluma hypogea (Porifera) is a carnivo- Electronic Supplementary Material Supplementary ma- rous species that belongs to the deep-sea taxon Cla- terial is available in the online version of this article at dorhizidae but lives in littoral caves and can be raised http://dx.doi.org/10.1007/s00435-004-0100-0 easily in an aquarium. It passively captures its prey by means of filaments covered with hook-like spicules. Various invertebrate species provided with setae or thin appendages are able to be captured, although minute crus- Introduction taceans up to 8 mm long are the most suitable prey. Multicellular animals almost universally feed by means of Transmission electron microscopy observations have been a digestive tract or a digestive cavity. Apart from some made during the digestion process. The prey is engulfed parasites directly living at the expense of their host, the in a few hours by the sponge cells, which migrate from only exceptions are the Pogonophores (deep-sea animals the whole body towards the prey and concentrate around relying on symbiotic chemoautotrophy and whose larvae it. A primary extracellular digestion possibly involving have a temporary digestive tract) and two groups of mi- the activity of sponge cells, autolysis of the prey and crophagous organisms relying on intracellular digestion, bacterial action results in the breaking down of the prey the minor Placozoa and, most importantly, sponges (Po- body. -

Lysosomes and the Connective Tissue Diseases LUCILLE BITENSKY from the Cellular Biology Division, Kennedy Institute of Rheumatology, Bute Gardens, London
J Clin Pathol: first published as 10.1136/jcp.31.Suppl_12.105 on 1 January 1978. Downloaded from J. clin. Path., 31, Suppl. (Roy. Coll. Path.), 12, 105-116 Lysosomes and the connective tissue diseases LUCILLE BITENSKY From the Cellular Biology Division, Kennedy Institute of Rheumatology, Bute Gardens, London Lysosomes are small intracellular organelles present respondingly, in phagocytic cells primary lysosomes in most or all cells of animals of widely different become attached to endocytic vacuoles and release evolutionary development. In general their diameter their hydrolytic enzymes into these vacuoles, digest- may vary from 0(2 to 0(5 ,tm so that they overlap the ing the endocytosed matter (Cohn and Fedorko, dimensions of mitochondria. In the original differen- 1969). tial centrifugation studies they were isolated as the 'light mitochondrial' fraction. It is difficult to give Methods for studying lysosomes an exact size to these organelles because the term 'Iysosome' covers a wide range of structures from the BIOCHEMICAL small primary lysosomes budded off from the Golgi The term 'Iysosome' was coined by de Duve (de apparatus, to secondary lysosomes caused by the Duve et al., 1955; de Duve, 1969) for particles fusion of primary lysosomes with endocytotic isolated by homogenisation and differential centri- vacuoles, to complex fusion-structures such as het- fugation that contained acid hydrolases in a latent erolysosomes and autophagic vacuoles. Indeed, form. The latency of the enzymic activities was the de Duve (1969) suggested that lysosomes are only a most surprising feature of this organelle. Now these part of the intracellular vacuolar system. Con- structures are usually separated from mitochondria by copyright. -

Brief Reports Enteroliths in a Meckel's Diverticulum Mimicking Gallstone
brief reports Enteroliths in a Meckel’s Diverticulum Mimicking Gallstone Ileus Peter G. Justus, MD, James J. Bergman, MD, and Thomas R. Reagan, MD Redmond, W ashington allstone ileus is a form of mechanical obstruction of pain. The recurring symptoms had a duration of several G the small intestine caused by a large gallstone that hours, each time culminating in severe distress lasting 10 has passed from the gallbladder to the intestine through to 15 minutes. Progressive fatigue and episodic anorexia acholecystoenteral fistula.1 This diagnosis is suspected in pursued. He denied fatty food intolerance, jaundice, vom patients with gallstones and symptoms or signs of inter iting, or a past history of gastrointestinal disease. The mittent small-bowel obstruction. family history and other personal history were unremark Meckel’s diverticulum, the most common malformation able. of the gastrointestinal tract, is found in about 2 percent On examination the patient was well nourished and in of individuals.2 Its rare clinical manifestations include in no acute distress. He was normotensive, afebrile, and testinal obstruction, ulceration, hemorrhage, intussus nonicteric. The abdomen was mildly distended, and there ception, and neoplasm. Such phenomena usually are not was mild tenderness to deep palpation of the right upper diagnosed preoperatively.3 Some 50 cases of enteroliths quadrant. No rebound or guarding was elicited, nor were occurring in a Meckel’s diverticulum have been reported.4 there masses noted. Rectal examination was negative, al This unusual finding occurs most often in men and can though some brown, guaiac-positive stool was recovered. present with lower abdominal pain, vomiting,5 rectal The remainder of the physical examination was within bleeding,6 anemia,7 or bowel obstruction.8 Radiographs normal limits. -

Jejunal Diverticula Causing a Life-Threatening Lower
Available online at www.ijmrhs.com cal R edi ese M ar of c l h a & n r H u e o a J l l t h International Journal of Medical Research & a S n ISSN No: 2319-5886 o c i t i Health Sciences, 2019, 8(4): 196-200 e a n n c r e e t s n I • • IJ M R H S Jejunal Diverticula Causing a Life-Threatening Lower Gastrointestinal Bleeding: A Case Report Abdelmoniem MM Makkawi* and Mohammed Eltoum Hamid Azoz Department of Surgery, University of Elimam Elmahadi, Kosti, Sudan *Corresponding e-mail: [email protected] ABSTRACT A jejunal diverticulum is a rare and usually asymptomatic disease. More commonly it is usually seen as incidental findings on radiological studies or during surgery. Complications such as bleeding, perforation, abscess formation, obstruction, malabsorption, blind loop syndrome, volvulus, and intussusception may warrant surgical intervention. Herein, we report a case of a 62-year old woman presenting with massive lower gastrointestinal bleeding, she was pale, clammy and hemodynamically unstable, she was initially resuscitated with IV fluids and whole blood, urgent upper endoscopy was normal, colonoscopy revealed sigmoid colon ulcerative lesion with histopathological evidence of adenocarcinoma, there was bleeding coming from upwards. After staging of the tumor, the decision was then made to proceed to exploratory laparotomy with a pre-operative plan of segmental colectomy. Intra-operatively segmental sigmoid colectomy was performed with end to end anastomosis, during formal laparotomy we found 2 giant diverticula in the proximal jejunum, small bowel resection and end to end anastomosis was done with the good postoperative outcome. -

Phylum Porifera
790 Chapter 28 | Invertebrates updated as new information is collected about the organisms of each phylum. 28.1 | Phylum Porifera By the end of this section, you will be able to do the following: • Describe the organizational features of the simplest multicellular organisms • Explain the various body forms and bodily functions of sponges As we have seen, the vast majority of invertebrate animals do not possess a defined bony vertebral endoskeleton, or a bony cranium. However, one of the most ancestral groups of deuterostome invertebrates, the Echinodermata, do produce tiny skeletal “bones” called ossicles that make up a true endoskeleton, or internal skeleton, covered by an epidermis. We will start our investigation with the simplest of all the invertebrates—animals sometimes classified within the clade Parazoa (“beside the animals”). This clade currently includes only the phylum Placozoa (containing a single species, Trichoplax adhaerens), and the phylum Porifera, containing the more familiar sponges (Figure 28.2). The split between the Parazoa and the Eumetazoa (all animal clades above Parazoa) likely took place over a billion years ago. We should reiterate here that the Porifera do not possess “true” tissues that are embryologically homologous to those of all other derived animal groups such as the insects and mammals. This is because they do not create a true gastrula during embryogenesis, and as a result do not produce a true endoderm or ectoderm. But even though they are not considered to have true tissues, they do have specialized cells that perform specific functions like tissues (for example, the external “pinacoderm” of a sponge acts like our epidermis). -

Zenker's Diverticulum and Squamous Esophageal Cancer
Journal of Mind and Medical Sciences Volume 4 | Issue 2 Article 15 2017 Zenker’s diverticulum and squamous esophageal cancer: a case report Ion Dina Carol Davila University, St. Ioan Clinical Hospital, Department of Gastroenterology, Bucharest, Romania Octav Ginghina Carol Davila University, St. Ioan Clinical Hospital, Department of Surgery, Bucharest, Romania Corina D. Toderescu Vasile Goldis Western University of Arad, Faculty of General Medicine, Arad, Romania Cristian Bălălău Carol Davila University, St. Pantelimon Hospital, Department of Surgery, Bucharest, Romania, [email protected] Bianca Galateanu University of Bucharest, Department of Biochemistry and Molecular Biology, Bucharest, Romania See next page for additional authors Follow this and additional works at: http://scholar.valpo.edu/jmms Part of the Medical Sciences Commons, and the Surgery Commons Recommended Citation Dina, Ion; Ginghina, Octav; Toderescu, Corina D.; Bălălău, Cristian; Galateanu, Bianca; Negrei, Carolina; and Iacobescu, Claudia (2017) "Zenker’s diverticulum and squamous esophageal cancer: a case report," Journal of Mind and Medical Sciences: Vol. 4 : Iss. 2 , Article 15. DOI: 10.22543/7674.42.P193197 Available at: http://scholar.valpo.edu/jmms/vol4/iss2/15 This Case Presentation is brought to you for free and open access by ValpoScholar. It has been accepted for inclusion in Journal of Mind and Medical Sciences by an authorized administrator of ValpoScholar. For more information, please contact a ValpoScholar staff member at [email protected]. Zenker’s diverticulum and squamous esophageal cancer: a case report Authors Ion Dina, Octav Ginghina, Corina D. Toderescu, Cristian Bălălău, Bianca Galateanu, Carolina Negrei, and Claudia Iacobescu This case presentation is available in Journal of Mind and Medical Sciences: http://scholar.valpo.edu/jmms/vol4/iss2/15 J Mind Med Sci. -

Feeding & Digestion
Feeding & Digestion Why eat? • Macronutrients Feeding & Digestion – Energy for all our metabolic processes – Monomers to build our polymers 1. Carbohydrates 2. Lipids 3. Protein • Micronutrients – Tiny amounts of vital elements & compounds our body cannot adequately synthesize 4. Vitamins 5. Minerals • Hydration 6. Water Niche: role played in a FEEDING community (grazer, predator, • “Eat”: Gr. -phagy; Lt. -vore scavenger, etc.) • Your food may not Some organisms have specialized niches so as to: • increase feeding efficiency wish to be eaten! • reduce competition • DIET & Optimal foraging model: Need to maximize benefits (energy/nutrients) while minimizing MORPHOLOGY costs (energy expended/risks) • FOOD CAPTURE • MECHANICAL PROCESSING Never give up! Optimal Foraging Model Optimal Foraging Model • Morphology reflects foraging strategies • Generalist is less limited by rarity of resources • Specialist is more efficient at exploiting a specific resource Generalist OMNIVORE opossum CARNIVORE HERBIVORE wolf elephant Optimal foraging strategies: Need to maximize benefits (energy/nutrients) INSECTIVORE PISCIVORE while minimizing costs (energy shrew osprey expended / risks) MYRMECOPHAGORE • Tapirs have 40x more meat, but are anteater much harder to find and catch. Specialist So jaguars prefer armadillos. Heyer 1 Feeding & Digestion Bird Beaks Anteater Adaptations • Generalist & specialist bills • Thick fur • Small eyes • Long claws in front • Long snout • Long barbed tongue • No teeth FOOD CAPTURE SPECIALIZATIONS Fluid Feeding – Fluid Feeding • Sucking with tube – Suspension & Deposit Feeding – mosquitoes & butterflies • Lapping with brushy tongue – Grazers & Browsers – hummingbirds, fruit bats – Predation: Ambush & Attraction – bees – Venoms – Tool Use & Team Efforts Hummingbird tongue Suspension Feeding (Filter Feeding) • Filter food (plankton, small animals, organic particles) suspended in water. • http://www.youtube.com/watch?v=1wpQ8HQEkvE • Filters are hard, soft or even sticky. -
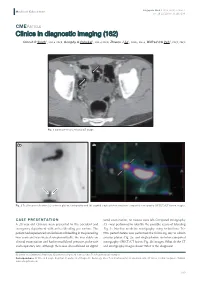
Clinics in Diagnostic Imaging (162)
Singapore Med J 2015; 56(9): 523-527 Medical Education doi: 10.11622/smedj.2015138 CMEARTICLE Clinics in diagnostic imaging (162) Dinesh R Singh1, MMed, FRCR, Geoiphy G Pulickal1, MMed, FRCR, Zhiwen J Lo2, MBBS, BMed, Wilfred CG Peh1, FRCP, FRCR Fig. 1 Contrast-enhanced axial CT image. 2a 2b Fig. 2 Tc-99m pertechnetate (a) anterior planar scintigraphy and (b) sagittal single-photon emission computed tomography (SPECT)/CT fusion images. CASE PRESENTATION rectal examination, no masses were felt. Computed tomography A 28-year-old Chinese man presented to the accident and (CT) was performed to identify the possible cause of bleeding emergency department with active bleeding per rectum. The (Fig. 1). Nuclear medicine scintigraphy using technetium (Tc)- patient had experienced similar bouts of bleeding in the preceding 99m pertechnetate was performed the following day to obtain two years and was treated symptomatically. He was stable on anterior planar (Fig. 2a) and single-photon emission computed clinical examination and had normal blood pressure, pulse rate tomography (SPECT)/CT fusion (Fig. 2b) images. What do the CT and respiratory rate. Although there was altered blood on digital and scintigraphy images show? What is the diagnosis? 1Department of Diagnostic Radiology, 2Department of General Surgery, Khoo Teck Puat Hospital, Singapore Correspondence: Dr Dinesh R Singh, Registrar, Department of Diagnostic Radiology, Khoo Teck Puat Hospital, Alexandra Health, 90 Yishun Central, Singapore 768828. [email protected] 523 Medical Education IMAGE INTERPRETATION Contrast-enhanced axial CT image (Fig. 1) shows posterior outpouching from a segment of the distal ileum (arrows) associated with mild wall thickening and increased mucosal enhancement. -

Intracellular Digestion
Intracellular Digestion By 0. ROGERANDERSON Downloaded from http://online.ucpress.edu/abt/article-pdf/32/8/461/27403/4443206.pdf by guest on 25 September 2021 It is now widely recognized that intracellular di- Thus intracellular digestion includes (i) the degrada- gestion is a process occurring in many different kinds tion of ingested food to produce energy-rich mole- of plant and animal cells. However, intracellular cules to support life-the process called phagotro- digestion is seldom discussed with much depth in phy; and (ii) the turnover of cellular structures to secondary-school biology classes. Some modern biol- provide reusable structural molecules in the syn- ogy curricula give only brief mention of it. Sometimes thesis of new organelles-the process called auto- the concept is presented as a specific protozoan pro- phagy. Cellular digestion of all kinds is accom- cess without further discussion of its general proper- plished, in part, by the digestive action of hydrolyt- ties. In view of the widespread occurrence of intra- ic enzymes called hydrolases. A list of hydrolases cellular digestion in living systems, secondary-school usually associated with cellular digestion is given, students should be introduced to the topic in its most along with their substrates, in the table. It should be general form. This paper (i) contains a discussion of obvious from examining this table that a wide range a general concept of intracellular digestion, (ii) pre- of substances found in living material can be de- sents evidence for its widespread occurrence in liv- graded by these enzymes. ing things, and (iii) makes some suggestions toward Extracellular digestion is known to occur in bac- application of these ideas in planning biology lessons. -

Giant Sigmoid Diverticulum: Clinical and Radiological Features
Gut: first published as 10.1136/gut.18.12.1051 on 1 December 1977. Downloaded from Gut, 1977, 18, 1051-1053 Giant sigmoid diverticulum: clinical and radiological features D. R. FOSTER AND B. ROSS1 From the Department of Radiology, Northern General Hospital, Sheffield SUMMARY Two case reports of giant sigmoid diverticulum associated with diverticular disease of the sigmoid colon are presented. The clinical and radiological features of 30 similar cases found in the literature are reviewed. Our two cases represent the largest recorded diverticulum and the oldest recorded patient with this condition. Diverticular disease of the colon is a common clinical finding. Its incidence on barium enema or post-mortem examination rises with increasing age so that at the age of 60 years it is found in at least 30 % of patients studied. Giant sigmoid diverticulum is a rare complication and only 30 cases have been described in the world literature since it was first reported by Hughes and Greene in 1953. Only one report of two cases (Johns and Hartley, 1976) has appeared in the British literature. Confirmation was obtained at operation in both our cases, which illustrate the typical radiological appearances. http://gut.bmj.com/ Case I A 61-year-old female was admitted with a three week history of central abdominal pain. This had become severe and persistent over the previous 12 hours and was accompanied by nausea and occasional vomiting. On clinical examination she appeared pale and unwell. There was considerable abdominal dis- on September 30, 2021 by guest. Protected copyright. tension but bowel sounds were normal.