Profiling of Human Lymphocytes Reveals a Specific Network of Protein Kinases Modulated by Endurance Training Status
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Supplementary Table 7. Characterization of Human Proteins Involved in the Prostate Cancer Pathway
Supplementary Table 7. Characterization of human proteins involved in the prostate cancer pathway f Protein UniProt Protein PONDR-FIT MobiDB Location (length) Location (length) Nint ID length (%)b consensus of long disordered of AIBSse a c d (NAIBS) (%) regions BAD, Bcl2-associated Q92934 168 100.00 84.54 1-105 (105) 1-53 (53) 66 agonist of cell death (4/70.8) 122-147 (27) 57-80 (24) 158-168 (11) 100-129 (30) 146-157 (12) CREB5; cyclic AMP- Q02930 508 85.24 75.39 46-59 (14) 66-86 (21) 65 responsive element (7/67.9) 86-393 (308) 99-183 (85) binding protein 5 447-470 (24) 188-358 (171) 479-508 (31) 362-370 (9) 378-406 (29) 421-444 (24) 503-508 (6) CREB1, cyclic AMP- P16220 341 79.47 40.47 1-32 (32) 32-44 (13) 169 responsive element- (7/29.3) 40-50 (11) 89-104 (16) binding protein 1 102-132 (33) 128-145 (18) 138-171 (34) 166-191 (26) 271-285 (15) 265-270 (6) 307-314 (8) 329-341 (13) FOXO1, Forkhead box Q12778 655 78.63 72.82 1-69 (69) 1-32 (32) 68 protein O1 (19/56.9) 74-101 (28) 54-82 (29) 105-160 (56) 88-118 (31) 199-210 (12) 160-172 (13) 229-336 (107) 182-196 (15) 385-450 (66) 216-226 (11) 463-488 (26) 258-280 (23) 498-569 (72) 289-297 (9) 644-655 (12) 306-314 (9) 323-365 (43) 371-388 (18) 301-409 (8) 447-469 (23) 483-517 (35) 528-545 (18) 550-565 (16) 570-592 (23) 605-612 (8) TCF7L1, transcription Q9HCS4 588 77.04 61.90 1-104 (104) 1-46 (46) 4 factor 7 like 1 (16/54.5) 161-183 (23) 53-74 (22) 192-238 (47) 94-135 (42) 316-344 (29) 146-159 (14) 406-512 (107) 191-201 (11) 524-546 (21) 234-252 (19) 274-288 (15) 349-371 (23) 373-383 (11) -

Molecular Classification of Patients with Unexplained Hamartomatous and Hyperplastic Polyposis
ORIGINAL CONTRIBUTION Molecular Classification of Patients With Unexplained Hamartomatous and Hyperplastic Polyposis Kevin Sweet, MS, CGC Context Significant proportions of patients with hamartomatous polyposis or with Joseph Willis, MD hyperplastic/mixed polyposis remain without specific clinical and molecular diagnosis Xiao-Ping Zhou, MD, PhD or present atypically. Assigning a syndromic diagnosis is important because it guides management, especially surveillance and prophylactic surgery. Carol Gallione, PhD Objective To systematically classify patients with unexplained hamartomatous or hy- Takeshi Sawada, MD, PhD perplastic/mixed polyposis by extensive molecular analysis in the context of central Pia Alhopuro, MD rereview of histopathology results. Sok Kean Khoo, PhD Design, Setting, and Patients Prospective, referral-based study of 49 unrelated patients from outside institutions (n=28) and at a comprehensive cancer center (n=21), Attila Patocs, MD, PhD conducted from May 2, 2002, until December 15, 2004. Germline analysis of PTEN, Cossette Martin, PhD BMPR1A, STK11 (sequence, deletion), SMAD4, and ENG (sequence), specific exon screen- Scott Bridgeman, BSc ing of BRAF, MYH, and BHD, and rereview of polyp histology results were performed. John Heinz, PhD Main Outcome Measures Molecular, clinical, and histopathological findings in pa- tients with unexplained polyposis. Robert Pilarski, MS, CGC Results Of the 49 patients, 11 (22%) had germline mutations. Of 14 patients with Rainer Lehtonen, BSc juvenile polyposis, 2 with early-onset disease had mutations in ENG, encoding endo- Thomas W. Prior, PhD glin, previously only associated with hereditary hemorrhagic telangiectasia; 1 had hemi- zygous deletion encompassing PTEN and BMPR1A; and 1 had an SMAD4 mutation. Thierry Frebourg, MD, PhD One individual previously classified with Peutz-Jeghers syndrome had a PTEN dele- Bin Tean Teh, MD, PhD tion. -

Inhibition of Mitochondrial Complex II in Neuronal Cells Triggers Unique
www.nature.com/scientificreports OPEN Inhibition of mitochondrial complex II in neuronal cells triggers unique pathways culminating in autophagy with implications for neurodegeneration Sathyanarayanan Ranganayaki1, Neema Jamshidi2, Mohamad Aiyaz3, Santhosh‑Kumar Rashmi4, Narayanappa Gayathri4, Pulleri Kandi Harsha5, Balasundaram Padmanabhan6 & Muchukunte Mukunda Srinivas Bharath7* Mitochondrial dysfunction and neurodegeneration underlie movement disorders such as Parkinson’s disease, Huntington’s disease and Manganism among others. As a corollary, inhibition of mitochondrial complex I (CI) and complex II (CII) by toxins 1‑methyl‑4‑phenylpyridinium (MPP+) and 3‑nitropropionic acid (3‑NPA) respectively, induced degenerative changes noted in such neurodegenerative diseases. We aimed to unravel the down‑stream pathways associated with CII inhibition and compared with CI inhibition and the Manganese (Mn) neurotoxicity. Genome‑wide transcriptomics of N27 neuronal cells exposed to 3‑NPA, compared with MPP+ and Mn revealed varied transcriptomic profle. Along with mitochondrial and synaptic pathways, Autophagy was the predominant pathway diferentially regulated in the 3‑NPA model with implications for neuronal survival. This pathway was unique to 3‑NPA, as substantiated by in silico modelling of the three toxins. Morphological and biochemical validation of autophagy markers in the cell model of 3‑NPA revealed incomplete autophagy mediated by mechanistic Target of Rapamycin Complex 2 (mTORC2) pathway. Interestingly, Brain Derived Neurotrophic Factor -

(12) United States Patent (10) Patent No.: US 7,863,289 B2 Spevak Et Al
US00786.3289B2 (12) United States Patent (10) Patent No.: US 7,863,289 B2 Spevak et al. (45) Date of Patent: *Jan. 4, 2011 (54) COMPOUNDS AND METHODS FOR KINASE 5,658,775 A 8, 1997 Gilboa MODULATION, AND INDICATIONS 5,681,959 A 10/1997 Bishop et al. THEREFOR 5,700,637 A 12/1997 Southern (75) Inventors: Wayne Spevak, Berkeley, CA (US); 5,700.809 A 12/1997 Leeson et al. Hanna Cho, Oakland, CA (US); Prabha 5,712,285 A 1/1998 Curtis et al. N. Ibrahim, Mountain View, CA (US); 5,721,118 A 2, 1998 Scheffler Shenghua Shi, San Diego, CA (US); 5,744,305 A 4/1998 Fodor et al. Shumeye Mamo, Oakland, CA (US); 5,747,276 A 5/1998 Hoch et al. Samuel J. Gillette, Oakland, CA (US); 5,763,198 A 6/1998 Hirth et al. Hongyao Zhu, Berkeley, CA (US) 5,770.456 A 6/1998 Holmes (73) Assignee: Plexxikon, Inc., Berkeley, CA (US) 5,800,992 A 9, 1998 Fodor et al. 5,807,522 A 9, 1998 Brown et al. (*) Notice: Subject to any disclaimer, the term of this 5,830,645 A 11/1998 Pinkel et al. patent is extended or adjusted under 35 5,840,485 A 11/1998 Leblet al. U.S.C. 154(b) by 356 days. 5,856,174 A 1/1999 Lipshutz et al. 5,877,007 A 3/1999 Housey This patent is Subject to a terminal dis claimer. 5,959,098 A 9/1999 Goldberg et al. 5,965,452 A 10, 1999 Kovacs (21) Appl. -

PDF Download
Tec Monoclonal Antibody Catalog No : YM0614 Reactivity : Human Applications : WB,ELISA Gene Name : TEC Protein Name : Tyrosine-protein kinase Tec Human Gene Id : 7006 Human Swiss Prot P42680 No : Mouse Swiss Prot P24604 No : Immunogen : Purified recombinant fragment of Tec expressed in E. Coli. Specificity : Tec Monoclonal Antibody detects endogenous levels of Tec protein. Formulation : Ascitic fluid containing 0.03% sodium azide,0.5% BSA, 50%glycerol. Source : Mouse Dilution : Western Blot: 1/500 - 1/2000. ELISA: 1/10000. Not yet tested in other applications. Purification : Affinity purification Storage Stability : -20°C/1 year Cell Pathway : T_Cell_Receptor, P References : 1. Blood. 2000 Nov 15;96(10):3406-13. 2. J Biol Chem. 1999 Jan 8;274(2):607-17. 3. Blood. 1998 Feb 1;91(3):940-8. Background : tec protein tyrosine kinase(TEC) Homo sapiens The protein encoded by this 1 / 2 gene belongs to the Tec family of non-receptor protein-tyrosine kinases containing a pleckstrin homology domain. Tec family kinases are involved in the intracellular signaling mechanisms of cytokine receptors, lymphocyte surface antigens, heterotrimeric G-protein coupled receptors, and integrin molecules. They are also key players in the regulation of the immune functions. Tec kinase is an integral component of T cell signaling and has a distinct role in T cell activation. This gene may be associated with myelodysplastic syndrome. [provided by RefSeq, Jul 2008], Function : catalytic activity:ATP + a [protein]-L-tyrosine = ADP + a [protein]-L-tyrosine phosphate.,caution:It is uncertain whether Met-1 is the initiator.,cofactor:Binds 1 zinc ion per subunit.,similarity:Belongs to the protein kinase superfamily. -

SNP Gene Chr* Region P Value Odd Ratios Minor Allele Major Allele Rs11184708 PRMT6 1 Upstream 6.447× 10−13 6.149 T a Rs108025
Supplementary Table S1. Detailed information on scrub typhus-related candidate SNPs with a p value < 1 × 10−4. Odd Minor Major SNP Gene Chr* Region p value Ratios Allele Allele rs11184708 PRMT6 1 upstream 6.447× 10−13 6.149 T A rs10802595 RYR2 1 intron 0.00008738 2.593 A G downstream, intron, rs401974 LINC00276,LOC100506474 2 0.0000769 0.3921 T C upstream rs1445126 MIR4757,NT5C1B 2 upstream 0.00004819 3.18 A G LOC101930107,MIR4435-1,PLGLB rs62140478 2 downstream, upstream 7.404 × 10−8 9.708 T C 2 rs35890165 CPS1,ERBB4 2 downstream 0.00003952 0.373 A G rs34599430 ZNF385D,ZNF385D-AS2 3 intron, upstream 0.00002317 2.767 G A rs6809058 RBMS3,TGFBR2 3 downstream, upstream 0.00002507 3.094 G A rs3773683 SIDT1 3 intron 0.00006635 0.3543 C T rs11727383 CRMP1,EVC 4 intron 0.0000803 0.3459 A G rs17338338 NUDT12,RAB9BP1 5 upstream 0.00006987 0.3746 G A rs2059950 DTWD2,LOC102467225 5 downstream 0.000008739 2.911 G A rs72663337 DTWD2,LOC102467225 5 downstream 0.00009856 2.566 T C rs6882516 LSM11 5 UTR-3 0.00007553 2.968 A C rs76949230 TENM2 5 intron 0.00006305 3.048 G C rs3804468 LY86,LY86-AS1 6 intron 0.00003155 0.202 C T rs3778337 DSP 6 exon,intron 0.00007311 0.3897 G A rs16883596 MAP3K7,MIR4643 6 upstream 0.00005186 4.274 A G rs35144103 CCT6P3,ZNF92 7 downstream, upstream 0.00004885 3.422 A G rs13244090 LOC407835,TPI1P2 7 downstream, upstream 0.00004505 2.638 G A rs17167553 LRGUK 7 missense 0.00008788 2.75 T G rs6583826 IDE,KIF11 10 upstream 0.00006894 0.2846 G A rs10769111 LOC221122,PRDM11 11 downstream, upstream 0.0000619 0.3816 T G rs10848921 -

Rabbit Anti-IKK Gamma/FITC Conjugated Antibody-SL2898R-FITC
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-IKK gamma/FITC Conjugated antibody SL2898R-FITC Product Name: Anti-IKK gamma/FITC Chinese Name: FITC标记的KB抑制蛋白激酶γ抗体 IkB kinase associated protein 1; IkB kinase subunit gamma; Inhibitor of nuclear factor kappa B kinase subunit gamma; AMCBX1; FIP 3; FIP3; Fip3p; I kappa B kinase gamma; IkB kinase associated protein 1; IkB kinase gamma subunit; IkB kinase subunit gamma; IKBKG; IKKAP 1; IKKAP1; IKKG; Incontinentia pigmenti; Inhibitor of kappa light polypeptide gene enhancer in B cells kinase gamma; Inhibitor of kappa light Alias: polypeptide gene enhancer in B cells, kinase gamma; Inhibitor of nuclear factor kappa B kinase gamma subunit; Inhibitor of nuclear factor kappa B kinase subunit gamma; IP 1; IP 2; IP; IP1; IP2; IPD2; NEMO; NF kappa B essential modifier; NF kappa B essential modulator; NF kappaB essential modifier; NF kappaB essential modulator; NFkappaB essential modulator. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat,Dog,Pig,Horse,Rabbit, IF=1:50-200 Applications: not yet tested in other applications. optimalwww.sunlongbiotech.com dilutions/concentrations should be determined by the end user. Molecular weight: 46kDa Form: Lyophilized or Liquid Concentration: 1mg/ml immunogen: KLH conjugated synthetic peptide derived from human IKK gamma Lsotype: IgG Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. -

Supplementary Information Supplementary Table 1
Supplementary information Supplementary Table 1 Kinase Type Format N Average SD (IC50) ALK Y Lantha 6 > 10 n.d. AURORA_A S/T Caliper 6 > 10 n.d. AXL Y Lantha 3 > 10 n.d. BTK Y Caliper 6 > 10 n.d. cABL Y Caliper 4 2.0 0.38 cABLT315 Y Caliper 6 > 10 n.d. CaMK2 S/T Caliper 3 > 10 n.d. CDK2A S/T Caliper 6 > 10 n.d. CDK4D1 S/T Caliper 3 > 10 n.d. CK1 S/T Caliper 3 > 10 n.d. cKIT Y Lantha 6 > 10 n.d. cMET Y Lantha 7 > 10 n.d. COT1 S/T Caliper 4 > 10 n.d. CSK Y Lantha 2 > 10 n.d. cSRC Y Lantha 3 > 10 n.d. EphA4 Y Lantha 6 > 10 n.d. EphB4 Y Lantha 6 > 10 n.d. ERK2 S/T Caliper 6 > 10 n.d. FAK Y Lantha 3 > 10 n.d. FGFR-1 Y Lantha 3 > 10 n.d. FGFR-2 Y Lantha 3 > 10 n.d. FGFR-3 Y Lantha 3 > 10 n.d. FGFR3K650E Y Lantha 6 > 10 n.d. FGFR-4 Y Lantha 6 > 10 n.d. FLT3 Y Lantha 3 > 10 n.d. FYN Y Lantha 3 > 10 n.d. GSK3beta S/T Caliper 6 > 10 n.d. HCK Y Lantha 3 > 10 n.d. HER1 Y Lantha 6 > 10 n.d. HER2 Y Lantha 6 > 10 n.d. HER4 Y Lantha 3 > 10 n.d. IGF1R Y Lantha 6 > 10 n.d. INS1R Y Lantha 6 > 10 n.d. IRAK4 S/T Caliper 3 > 10 n.d. -
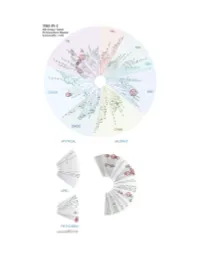
Profiling Data
Entrez Gene Percent Compound Compound Name DiscoveRx Gene Symbol Symbol Control Concentration (nM) THZ-P1-2 AAK1 AAK1 100 1000 THZ-P1-2 ABL1(E255K)-phosphorylated ABL1 21 1000 THZ-P1-2 ABL1(F317I)-nonphosphorylated ABL1 93 1000 THZ-P1-2 ABL1(F317I)-phosphorylated ABL1 100 1000 THZ-P1-2 ABL1(F317L)-nonphosphorylated ABL1 71 1000 THZ-P1-2 ABL1(F317L)-phosphorylated ABL1 44 1000 THZ-P1-2 ABL1(H396P)-nonphosphorylated ABL1 5.8 1000 THZ-P1-2 ABL1(H396P)-phosphorylated ABL1 6.6 1000 THZ-P1-2 ABL1(M351T)-phosphorylated ABL1 12 1000 THZ-P1-2 ABL1(Q252H)-nonphosphorylated ABL1 24 1000 THZ-P1-2 ABL1(Q252H)-phosphorylated ABL1 20 1000 THZ-P1-2 ABL1(T315I)-nonphosphorylated ABL1 93 1000 THZ-P1-2 ABL1(T315I)-phosphorylated ABL1 100 1000 THZ-P1-2 ABL1(Y253F)-phosphorylated ABL1 2.4 1000 THZ-P1-2 ABL1-nonphosphorylated ABL1 13 1000 THZ-P1-2 ABL1-phosphorylated ABL1 8.1 1000 THZ-P1-2 ABL2 ABL2 36 1000 THZ-P1-2 ACVR1 ACVR1 94 1000 THZ-P1-2 ACVR1B ACVR1B 100 1000 THZ-P1-2 ACVR2A ACVR2A 94 1000 THZ-P1-2 ACVR2B ACVR2B 91 1000 THZ-P1-2 ACVRL1 ACVRL1 90 1000 THZ-P1-2 ADCK3 CABC1 77 1000 THZ-P1-2 ADCK4 ADCK4 97 1000 THZ-P1-2 AKT1 AKT1 95 1000 THZ-P1-2 AKT2 AKT2 95 1000 THZ-P1-2 AKT3 AKT3 100 1000 THZ-P1-2 ALK ALK 92 1000 THZ-P1-2 ALK(C1156Y) ALK 93 1000 THZ-P1-2 ALK(L1196M) ALK 71 1000 THZ-P1-2 AMPK-alpha1 PRKAA1 93 1000 THZ-P1-2 AMPK-alpha2 PRKAA2 100 1000 THZ-P1-2 ANKK1 ANKK1 97 1000 THZ-P1-2 ARK5 NUAK1 83 1000 THZ-P1-2 ASK1 MAP3K5 100 1000 THZ-P1-2 ASK2 MAP3K6 95 1000 THZ-P1-2 AURKA AURKA 99 1000 THZ-P1-2 AURKB AURKB 100 1000 THZ-P1-2 AURKC AURKC 83 1000 -

LANCE Ultra Kinase Assay Selection Guide
FINDING THE PATHWAY TO ASSAY OPTIMIZATION IS EASY LANCE® Ultra Kinase Assay Selection Guide LANCE Ultra Serine/Threonine Kinase Selection Guide LANCE® Ultra TR-FRET reagents comprise the widest portfolio of validated kinase assay offerings available for rapid, sensitive and robust screening of purified kinase targets in a biochemical format. • We provide S/B ratiometric data for each LANCE Ultra assay to guide you to • Our selection guides contain over 300 kinases from a variety of suppliers: the best performing solution for your assay. – 225 Serine/Threonine kinases validated on LANCE Ultra reagents • Rapid assay optimization every time. – 85 Tyrosine kinases validated on LANCE Ultra reagents How to use this guide: 1. Locate your kinase If you cannot find your kinase of interest, please ask your PerkinElmer sales • In many cases, up to three commercial kinase vendors have been tested. specialist, as our list continues to expand. Two kits are available for testing purposes: • Many common aliases are shown in parenthesis. • KinaSelect Ser/Thr kit (5 x 250 data points, TRF0300-C) 2. Best performing ULight ™ substrates are listed for each enzyme according to performance – 5 ULight-labeled Ser/Thr kinase specific substrates + 5 matching Europium-labeled anti-phospho antibodies • Signal to background (S/B) ratios (Signal at 665 nm / minus ATP control at 665 nm) are indicated in parenthesis. • KinaSelect TK kit (1,000 data points, TRF0301-D) • All S/B ratios were obtained at fixed experimental conditions unless – 1 ULight-labeled kinase specific substrate + 1 matching otherwise noted (see page 10). Europium-labeled anti-phospho antibody 3. Based on your substrate choice, find the corresponding Europium-labeled anti-phospho antibody on page 11 (i.e. -

A Nonsense Mutation in the IKBKG Gene in Mares with Incontinentia Pigmenti
A Nonsense Mutation in the IKBKG Gene in Mares with Incontinentia Pigmenti Rachel E. Towers1,2., Leonardo Murgiano3,4., David S. Millar1, Elise Glen2, Ana Topf2, Vidhya Jagannathan3,4, Cord Dro¨ gemu¨ ller3,4, Judith A. Goodship2, Angus J. Clarke1, Tosso Leeb3,4* 1 Institute of Medical Genetics, Cardiff University, Cardiff, United Kingdom, 2 Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, United Kingdom, 3 Institute of Genetics, Vetsuisse Faculty, University of Bern, Bern, Switzerland, 4 DermFocus, Vetsuisse Faculty, University of Bern, Bern, Switzerland Abstract Ectodermal dysplasias (EDs) are a large and heterogeneous group of hereditary disorders characterized by abnormalities in structures of ectodermal origin. Incontinentia pigmenti (IP) is an ED characterized by skin lesions evolving over time, as well as dental, nail, and ocular abnormalities. Due to X-linked dominant inheritance IP symptoms can only be seen in female individuals while affected males die during development in utero. We observed a family of horses, in which several mares developed signs of a skin disorder reminiscent of human IP. Cutaneous manifestations in affected horses included the development of pruritic, exudative lesions soon after birth. These developed into wart-like lesions and areas of alopecia with occasional wooly hair re-growth. Affected horses also had streaks of darker and lighter coat coloration from birth. The observation that only females were affected together with a high number of spontaneous abortions suggested an X-linked dominant mechanism of transmission. Using next generation sequencing we sequenced the whole genome of one affected mare. We analyzed the sequence data for non-synonymous variants in candidate genes and found a heterozygous nonsense variant in the X-chromosomal IKBKG gene (c.184C.T; p.Arg62*).