Automatic Mass Detection in Breast Using Deep Convolutional Neural Network and SVM Classifier Md
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BREAST IMAGING for SCREENING and DIAGNOSING CANCER Policy Number: DIAGNOSTIC 105.9 T2 Effective Date: January 1, 2017
Oxford UnitedHealthcare® Oxford Clinical Policy BREAST IMAGING FOR SCREENING AND DIAGNOSING CANCER Policy Number: DIAGNOSTIC 105.9 T2 Effective Date: January 1, 2017 Table of Contents Page Related Policies INSTRUCTIONS FOR USE .......................................... 1 Omnibus Codes CONDITIONS OF COVERAGE ...................................... 1 Preventive Care Services BENEFIT CONSIDERATIONS ...................................... 2 Radiology Procedures Requiring Precertification for COVERAGE RATIONALE ............................................. 3 eviCore Healthcare Arrangement APPLICABLE CODES ................................................. 5 DESCRIPTION OF SERVICES ...................................... 6 CLINICAL EVIDENCE ................................................. 7 U.S. FOOD AND DRUG ADMINISTRATION ................... 16 REFERENCES .......................................................... 18 POLICY HISTORY/REVISION INFORMATION ................ 22 INSTRUCTIONS FOR USE This Clinical Policy provides assistance in interpreting Oxford benefit plans. Unless otherwise stated, Oxford policies do not apply to Medicare Advantage members. Oxford reserves the right, in its sole discretion, to modify its policies as necessary. This Clinical Policy is provided for informational purposes. It does not constitute medical advice. The term Oxford includes Oxford Health Plans, LLC and all of its subsidiaries as appropriate for these policies. When deciding coverage, the member specific benefit plan document must be referenced. The terms -

Breast Scintimammography
CLINICAL MEDICAL POLICY Policy Name: Breast Scintimammography Policy Number: MP-105-MD-PA Responsible Department(s): Medical Management Provider Notice Date: 11/23/2020 Issue Date: 11/23/2020 Effective Date: 12/21/2020 Next Annual Review: 10/2021 Revision Date: 09/16/2020 Products: Gateway Health℠ Medicaid Application: All participating hospitals and providers Page Number(s): 1 of 5 DISCLAIMER Gateway Health℠ (Gateway) medical policy is intended to serve only as a general reference resource regarding coverage for the services described. This policy does not constitute medical advice and is not intended to govern or otherwise influence medical decisions. POLICY STATEMENT Gateway Health℠ does not provide coverage in the Company’s Medicaid products for breast scintimammography. The service is considered experimental and investigational in all applications, including but not limited to use as an adjunct to mammography or in staging the axillary lymph nodes. This policy is designed to address medical necessity guidelines that are appropriate for the majority of individuals with a particular disease, illness or condition. Each person’s unique clinical circumstances warrant individual consideration, based upon review of applicable medical records. (Current applicable Pennsylvania HealthChoices Agreement Section V. Program Requirements, B. Prior Authorization of Services, 1. General Prior Authorization Requirements.) Policy No. MP-105-MD-PA Page 1 of 5 DEFINITIONS Prior Authorization Review Panel – A panel of representatives from within the Pennsylvania Department of Human Services who have been assigned organizational responsibility for the review, approval and denial of all PH-MCO Prior Authorization policies and procedures. Scintimammography A noninvasive supplemental diagnostic testing technology that requires the use of radiopharmaceuticals in order to detect tissues within the breast that accumulate higher levels of radioactive tracer that emit gamma radiation. -

Evicore Breast Imaging Guidelines
CLINICAL GUIDELINES Breast Imaging Guidelines Version 2.0 Effective September 1, 2021 eviCore healthcare Clinical Decision Support Tool Diagnostic Strategies: This tool addresses common symptoms and symptom complexes. Imaging requests for individuals with atypical symptoms or clinical presentations that are not specifically addressed will require physician review. Consultation with the referring physician, specialist and/or individual’s Primary Care Physician (PCP) may provide additional insight. CPT® (Current Procedural Terminology) is a registered trademark of the American Medical Association (AMA). CPT® five digit codes, nomenclature and other data are copyright 2017 American Medical Association. All Rights Reserved. No fee schedules, basic units, relative values or related listings are included in the CPT® book. AMA does not directly or indirectly practice medicine or dispense medical services. AMA assumes no liability for the data contained herein or not contained herein. © 2021 eviCore healthcare. All rights reserved. Breast Imaging Guidelines V2.0 Breast Imaging Guidelines Abbreviations for Breast Guidelines 3 BR-Preface1: General Considerations 5 BR-1: Breast Ultrasound 8 BR-2: MRI Breast 10 BR-3: Breast Reconstruction 12 BR-4: MRI Breast is NOT Indicated 13 BR-5: MRI Breast Indications 14 BR-6: Nipple Discharge/Galactorrhea 18 BR-7: Breast Pain (Mastodynia) 19 BR-8: Alternative Breast Imaging Approaches 20 BR-9: Suspected Breast Cancer in Males 22 BR-10: Breast Evaluation in Pregnant or Lactating Females 23 ______________________________________________________________________________________________________ -

Procedure Guideline for Breast Scintigraphy
Procedure Guideline for Breast Scintigraphy Iraj Khalkhali, Linda E. Diggles, Raymond Taillefer, Penny R. Vandestreek, Patrick J. Peller and Hani H. Abdel-Nabi Harbor-UCLA Medical Center, Terranee; Nuclear Imaging Consultants, Roseville, California; Hospital Hôtel-Dieu de Montreal, Montreal, Quebec, Canada; Lutheran General Hospital, Park Ridge, Illinois; and University of Buffalo, Buffalo, New York Key Words: breast scintigraphy;procedureguideline should be available, as well as sonograms, if J NucÃMed 1999; 40:1233-1235 obtained. 2. A breast physical examination must be performed by either the nuclear medicine physician or the PART I: PURPOSE referring physician. 3. The time of last menses and pregnancy and lactat- The purpose of this guideline is to assist nuclear medicine ing status of the patient should be determined. practitioners in recommending, performing, interpreting and reporting the results of 99mTc-sestamibi breast scintigraphy 4. Breast scintigraphy should be delayed at least 2 wk after cyst or fine-needle aspiration, and 4—6wk (mammoscintigraphy, scintimammography). after core or excisional biopsy. 5. The nuclear medicine physician should be aware of PART II: BACKGROUND INFORMATION AND DEFINITIONS physical signs and symptoms and prior surgical procedures or therapy. Breast scintigraphy is performed after intravenous admin istration of "mTc-sestamibi and includes planar and/or C. Precautions None SPECT. D. Radiopharmaceutical 1. Intravenous injection of 740-1110 MBq (20-30 PART III: COMMON INDICATIONS AND APPLICATIONS mCi) 99mTc-sestamibi should be administered in an A. Evaluate breast cancer in patients in whom mammog- arm vein contralateral to the breast with the sus raphy is not diagnostic or is difficult to interpret (e.g., pected abnormality. -

Biopsy Needles and Drainage Catheters
Needles & Accessories | Catheters & Accessories Dilation & Accessories Spinal & Accessories | Implantable & Accessories Product Catalog ISO 13485 & ISO 9001 Certified Company Rev. 01 - 2019/03 About ADRIA Srl. Adria is a worldwide leader in developing, manufacturing and marketing healthcare products. The main focus is on Radiology, Oncology, Urology, Gynecology and Surgery . Adria' s corporate headquarter is based in Italy, it is ISO Certified and products are CE . marked. Adria was incorporated more than 20 years ago in Bologna , where the corporate headquarter and production plant is located. Adria is leader in developing and manufacturing healthcare products and keeps the status to be one of the first companies aimed to develop single patient use biopsy needles and drainage catheters. Over the time, thanks to the experience of specialized doctors and engineers , Adria product range and quality have been progressively enhanced, involving and developing the spine treatment line. Nowadays Adria has a worldwide presence . Reference markets are France, Spain, Turkey and products are distributed in more than 50 countries, through a large and qualified network of dealers. Since far off the very beginning, many things have changed, but Adria' s philosophy and purpose have always remained unchanged: helping healthcare providers to fulfill their mission of caring for patients. Table of Contents Needles & Accessories …………………………………………….....…...3 Catheters & Accessories ……………………….………………..…...…...18 Dilation & Accessories ……………………………………………...…...25 Spinal & Accessories ……………………………………………...…...30 Implantables & Accessories……………………………………………...…...35 Needles & Accessories HYSTO SYSTEM Automatic Biopsy Instrument……………………….……….…….. 4 HYSTO SYSTEM II Automatic Biopsy Instrument…………………….……….……...5 SAMPLE MASTER Semi-Automatic Biopsy Instrument…………………………….. 6 HYSTO-ONE Automatic Reusable Biopsy Instrument & MDA Biopsy Needle ....... 7 HYSTO-TWO Automatic Reusable Biopsy Instrument & MDS Biopsy Needle…... -

A Molecular Approach to Breast Imaging
Journal of Nuclear Medicine, published on January 16, 2014 as doi:10.2967/jnumed.113.126102 FOCUS ON MOLECULAR IMAGING A Molecular Approach to Breast Imaging Amy M. Fowler Department of Radiology, University of Wisconsin–Madison, Madison, Wisconsin malignant cells. A recent meta-analysis of the accuracy of 99mTc-sestamibi scintimammography as an adjunct to di- Molecular imaging is a multimodality discipline for noninvasively agnostic mammography for detection of breast cancer dem- visualizing biologic processes at the subcellular level. Clinical applications of radionuclide-based molecular imaging for breast onstrated a sensitivity of 83% and specificity of 85% (2). cancer continue to evolve. Whole-body imaging, with scinti- However, sensitivity was less for nonpalpable (59%) versus mammography and PET, and newer dedicated breast imaging palpable lesions (87%) despite comparable specificity, with systems are reviewed. The potential clinical indications and the no significant difference between planar and SPECT meth- challenges of implementing these emerging technologies are ods. Decreased sensitivity for nonpalpable, presumably presented. smaller, lesions is in part due to the limited spatial resolu- Key Words: molecular imaging; oncology; breast; PET; PET/ tion of conventional g cameras. CT; radiopharmaceuticals; breast cancer; breast-specific g im- In addition to 99mTc-sestamibi, the positron-emitting ra- aging; positron-emission mammography; positron-emission to- diopharmaceutical 18F-FDG accumulates in many types of mography cancer including breast. Meta-analyses of the accuracy of J Nucl Med 2014; 55:1–4 whole-body 18F-FDG PET used after standard diagnostic DOI: 10.2967/jnumed.113.126102 workup for patients with suspected breast lesions demon- strated sensitivities of 83%–89% and specificities of 74%– 80% (3,4). -

Evaluation of Nipple Discharge
New 2016 American College of Radiology ACR Appropriateness Criteria® Evaluation of Nipple Discharge Variant 1: Physiologic nipple discharge. Female of any age. Initial imaging examination. Radiologic Procedure Rating Comments RRL* Mammography diagnostic 1 See references [2,4-7]. ☢☢ Digital breast tomosynthesis diagnostic 1 See references [2,4-7]. ☢☢ US breast 1 See references [2,4-7]. O MRI breast without and with IV contrast 1 See references [2,4-7]. O MRI breast without IV contrast 1 See references [2,4-7]. O FDG-PEM 1 See references [2,4-7]. ☢☢☢☢ Sestamibi MBI 1 See references [2,4-7]. ☢☢☢ Ductography 1 See references [2,4-7]. ☢☢ Image-guided core biopsy breast 1 See references [2,4-7]. Varies Image-guided fine needle aspiration breast 1 Varies *Relative Rating Scale: 1,2,3 Usually not appropriate; 4,5,6 May be appropriate; 7,8,9 Usually appropriate Radiation Level Variant 2: Pathologic nipple discharge. Male or female 40 years of age or older. Initial imaging examination. Radiologic Procedure Rating Comments RRL* See references [3,6,8,10,13,14,16,25- Mammography diagnostic 9 29,32,34,42-44,71-73]. ☢☢ See references [3,6,8,10,13,14,16,25- Digital breast tomosynthesis diagnostic 9 29,32,34,42-44,71-73]. ☢☢ US is usually complementary to mammography. It can be an alternative to mammography if the patient had a recent US breast 9 mammogram or is pregnant. See O references [3,5,10,12,13,16,25,30,31,45- 49]. MRI breast without and with IV contrast 1 See references [3,8,23,24,35,46,51-55]. -

Evaluation of the Quantitative Accuracy of a Commercially-Available Positron Emission Mammography Scanner
The Texas Medical Center Library DigitalCommons@TMC The University of Texas MD Anderson Cancer Center UTHealth Graduate School of The University of Texas MD Anderson Cancer Biomedical Sciences Dissertations and Theses Center UTHealth Graduate School of (Open Access) Biomedical Sciences 8-2010 EVALUATION OF THE QUANTITATIVE ACCURACY OF A COMMERCIALLY-AVAILABLE POSITRON EMISSION MAMMOGRAPHY SCANNER Adam Springer Follow this and additional works at: https://digitalcommons.library.tmc.edu/utgsbs_dissertations Part of the Diagnosis Commons, Equipment and Supplies Commons, and the Other Medical Sciences Commons Recommended Citation Springer, Adam, "EVALUATION OF THE QUANTITATIVE ACCURACY OF A COMMERCIALLY-AVAILABLE POSITRON EMISSION MAMMOGRAPHY SCANNER" (2010). The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access). 64. https://digitalcommons.library.tmc.edu/utgsbs_dissertations/64 This Thesis (MS) is brought to you for free and open access by the The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences at DigitalCommons@TMC. It has been accepted for inclusion in The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences Dissertations and Theses (Open Access) by an authorized administrator of DigitalCommons@TMC. For more information, please contact [email protected]. EVALUATION OF THE QUANTITATIVE ACCURACY OF A COMMERCIALLY- AVAILABLE POSITRON EMISSION MAMMOGRAPHY SCANNER -
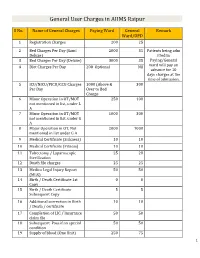
General User Charges in AIIMS Raipur
General User Charges in AIIMS Raipur S No. Name of General Charges Paying Ward General Remark Ward/OPD 1 Registration Charges 200 25 2 Bed Charges Per Day (Sami 2000 35 Patients being adm Deluxe) itted in 3 Bed Charges Per Day (Deluxe) 3000 35 Paying/General 4 Diet Charges Per Day 200 Optional Nil ward will pay an advance for 10 days charges at the time of admission. 5 ICU/NICU/PICU/CCU Charges 1000 (Above & 300 Per Day Over to Bed Charge 6 Minor Operation in OT/MOT 250 100 not mentioned in list, under L A 7 Minor Operation in OT/MOT 1000 300 not mentioned in list, under G A 8 Major Operation in OT, Not 2000 1000 mentioned in list under G A 9 Medical Certificate (Sickness) 10 10 10 Medical Certificate (Fitness) 10 10 11 Tubectomy / Laparoscopic 25 20 Sterilization 12 Death file charges 25 25 13 Medico Legal Injury Report 50 50 (MLR) 14 Birth / Death Certificate 1st 0 0 Copy 15 Birth / Death Certificate 5 5 Subsequent Copy 16 Additional correction in Birth 10 10 / Death / certificate 17 Completion of LIC / Insurance 50 50 claim file 18 Subsequent Pass if on special 50 50 condition 19 Supply of blood (One Unit) 250 75 1 20 Medical Board Certificate 500 500 On Special Case User Charges for Investigations in AIIMS Raipur S No. Name of Investigations Paying General Remark Ward Ward/OPD Anaesthsia 1 ABG 75 50 2 ABG ALONGWITH 150 100 ELECTROLYTES(NA+,K+)(Na,K) 3 ONLY ELECTROLYTES(Na+,K+,Cl,Ca+) 75 50 4 ONLY CALCIUM 50 25 5 GLUCOSE 25 20 6 LACTATE 25 20 7 UREA. -

Breast Self-Examination Practices in Women Who
Breast self-examination practices in women who have had a mastectomy by Carole H Crowell A thesis submitted in partial fulfillment of the requirement for the degree of Master of Nursing Montana State University © Copyright by Carole H Crowell (1990) Abstract: Research findings indicate that breast cancer mortality has not decreased significantly in the past decade. While research continues on the development of more effective therapies, techniques for early detection are presently receiving greater attention. The use of breast self-examination (BSE) has been emphasized in the media and professional literature but little emphasis has been placed on BSE for women who have had a mastectomy. The purposes of this study were to determine if women who have had breast surgery do breast self-examination and to describe barriers that block this behavior. An assumption was made that women who have had surgery for breast cancer do not examine their remaining breast tissue and scar site. A qualitative study was conducted using a grounded theory approach to explore and describe behavior of women who have had a mastectomy. Analytic strategies used for data analysis were the constant comparative method, theoretical sampling, open coding, memo writing, and recoding. A convenience sample of twelve informants was selected from a mastectomy support group located in a rural community in Montana. BSE was viewed as a health-promoting behavior and was found to relate to certain variables of Penders (1987) Health Promotion Model. Findings of this study indicate a majority of women perform BSE after their mastectomy but most do not examine their scar site. -

5. Effectiveness of Breast Cancer Screening
5. EFFECTIVENESS OF BREAST CANCER SCREENING This section considers measures of screening Nevertheless, the performance of a screening quality and major beneficial and harmful programme should be monitored to identify and outcomes. Beneficial outcomes include reduc- remedy shortcomings before enough time has tions in deaths from breast cancer and in elapsed to enable observation of mortality effects. advanced-stage disease, and the main example of a harmful outcome is overdiagnosis of breast (a) Screening standards cancer. The absolute reduction in breast cancer The randomized trials performed during mortality achieved by a particular screening the past 30 years have enabled the suggestion programme is the most crucial indicator of of several indicators of quality assurance for a programme’s effectiveness. This may vary screening services (Day et al., 1989; Tabár et according to the risk of breast cancer death in al., 1992; Feig, 2007; Perry et al., 2008; Wilson the target population, the rate of participation & Liston, 2011), including screening participa- in screening programmes, and the time scale tion rates, rates of recall for assessment, rates observed (Duffy et al., 2013). The technical quality of percutaneous and surgical biopsy, and breast of the screening, in both radiographic and radio- cancer detection rates. Detection rates are often logical terms, also has an impact on breast cancer classified by invasive/in situ status, tumour size, mortality. The observational analysis of breast lymph-node status, and histological grade. cancer mortality and of a screening programme’s Table 5.1 and Table 5.2 show selected quality performance may be assessed against several standards developed in England by the National process indicators. -

Ultrasound, Elastography and MRI Mammography
EAS Journal of Radiology and Imaging Technology Abbreviated Key Title: EAS J Radiol Imaging Technol ISSN 2663-1008 (Print) & ISSN: 2663-7340 (Online) Published By East African Scholars Publisher, Kenya Volume-1 | Issue-2 | Mar-Apr-2019 | Research Article Ultrasound, Elastography and MRI Mammography Correlation in Breast Pathologies (A Study of 50 Cases) Dr Hiral Parekh.1, Dr Lata Kumari.2, Dr Dharmesh Vasavada.3 1Professor, Department of Radiodiagnosis M P Shah Government Medical College Jamnagar, Gujarat, India 2Resident Doctor in Radiodiagnosis Department of Radiodiagnosis M P Shah Government Medical College Jamnagar, Gujarat, India 3Professor, Department of Surgery M P Shah Government Medical College Jamnagar, Gujarat, India *Corresponding Author Dr Dharmesh Vasavada Abstract: Introduction: The purpose of this study is to investigate the value of MRI in comparison to US and mammography in diagnosis of breast lesions. MRI is ideal for breast imaging due to its ability to depict excellent soft tissue contrast. Methods: This study of 50 cases was conducted in the department of Radiodiagnosis, Guru Gobinsingh Government Hospital, M P Shah Government Medical College, Jamnagar, Gujarat, India. All 50 cases having or suspected to have breast lesions were chosen at random among the indoor and outdoor patients referred to the Department of Radiodiagnosis for imaging. Discussion: In the present study the results of sonoelastography were compared with MRI. The malignant masses were the commonest and the mean age of patients with malignant masses in our study was 45 years, which is in consistent with Park‟s statement that the mean age of breast cancer occurrence is about 42 years in India3.