Clones in the Chick Diencephalon Contain Multiple Cell Types and Siblings Are Widely Dispersed
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Lecture 12 Notes
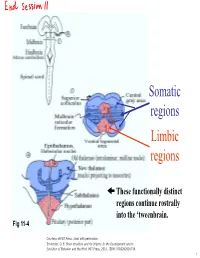
Somatic regions Limbic regions These functionally distinct regions continue rostrally into the ‘tweenbrain. Fig 11-4 Courtesy of MIT Press. Used with permission. Schneider, G. E. Brain structure and its Origins: In the Development and in Evolution of Behavior and the Mind. MIT Press, 2014. ISBN: 9780262026734. 1 Chapter 11, questions about the somatic regions: 4) There are motor neurons located in the midbrain. What movements do those motor neurons control? (These direct outputs of the midbrain are not a subject of much discussion in the chapter.) 5) At the base of the midbrain (ventral side) one finds a fiber bundle that shows great differences in relative size in different species. Give examples. What are the fibers called and where do they originate? 8) A decussating group of axons called the brachium conjunctivum also varies greatly in size in different species. It is largest in species with the largest neocortex but does not come from the neocortex. From which structure does it come? Where does it terminate? (Try to guess before you look it up.) 2 Motor neurons of the midbrain that control somatic muscles: the oculomotor nuclei of cranial nerves III and IV. At this level, the oculomotor nucleus of nerve III is present. Fibers from retina to Superior Colliculus Brachium of Inferior Colliculus (auditory pathway to thalamus, also to SC) Oculomotor nucleus Spinothalamic tract (somatosensory; some fibers terminate in SC) Medial lemniscus Cerebral peduncle: contains Red corticospinal + corticopontine fibers, + cortex to hindbrain fibers nucleus (n. ruber) Tectospinal tract Rubrospinal tract Courtesy of MIT Press. Used with permission. Schneider, G. -

Diencephalon Sists of the Midbrain, Pons, and Medulla
Diencephalon The brain lies within the cranial cavity of the skull and is made up of billions of nerve cells (neu - rons) and supporting cells (glia) . Neuronal cell bodies group together as gray matter, and their processes group together as white matter. The brain can be divided into four main parts: the cerebrum, diencephalon, brain stem, and cere - bellum. • The cerebrum is the largest part of the brain and consists of the four paired lobes with the two cerebral hemispheres, connected by a mass of white matter called the corpus callosum. The cerebrum accounts for about 80% of the brain’s mass and is concerned with higher functions, including perception of sensory impulses, instigation of voluntary movement, memory, thought, and reasoning. There are two layers of the cerebrum: - The cerebral cortex is the thin, wrinkled gray matter covering each hemisphere - The cerebral medulla is a thicker core of white matter • The diencephalon lies beneath the cerebral hemispheres and has two main structures ¾ the thalamus and the hypothalamus. The walnut-sized thalamus is a large mass of gray matter that lies on either side of the third ventricle. The thalamus is a great relay station on the afferent sen - sory pathway to the cerebral cortex. The tiny hypothalamus forms the lower part of the lateral wall and floor of the third ventricle. The hypothalamus exerts an influence on a wide range of body functions. • The cerebellum is attached to the brain and features a highly folded surface. It is important in the control of movement and balance. • The brainstem is the lower extension of the brain where it connects to the spinal cord. -

Chapter 128: Basic Mechanisms of Sleep: New Evidence On
128 BASIC MECHANISMS OF SLEEP: NEW EVIDENCE ON THE NEUROANATOMY AND NEUROMODULATION OF THE NREM-REM CYCLE EDWARD F. PACE-SCHOTT J. ALLAN HOBSON The 1990s brought a wealth of new detail to our knowledge NREM sleep (noradrenergic, serotonergic, and cholinergic of the brain structures involved in the control of sleep and systems damped), and REM sleep (noradrenergic and sero- waking and in the cellular level mechanisms that orchestrate tonergic systems off, cholinergic system undamped) (1–4). the sleep cycle through neuromodulation. This chapter pre- sents these new findings in the context of the general history Original Reciprocal Interaction Model: An of research on the brainstem neuromodulatory systems and Aminergic-Cholinergic Interplay the more specific organization of those systems in the con- trol of the alternation of wake, non–rapid eye movement The model of reciprocal interaction (5) provided a theoretic (NREM), and REM sleep. framework for experimental interventions at the cellular and Although the main focus of the chapter is on the our molecular level that has vindicated the notion that waking own model of reciprocal aminergic-cholinergic interaction, and REM sleep are at opposite ends of an aminergically we review new data suggesting the involvement of many dominant to cholinergically dominant neuromodulatory more chemically specific neuronal groups than can be ac- continuum, with NREM sleep holding an intermediate po- commodated by that model. We also extend our purview to sition (Fig. 128.1). The reciprocal interaction hypothesis the way in which the brainstem interacts with the forebrain. (5) provided a description of the aminergic-cholinergic in- These considerations inform not only sleep-cycle control terplay at the synaptic level and a mathematic analysis of per se, but also the way that circadian and ultradian rhythms the dynamics of the neurobiological control system. -

Brain Anatomy
BRAIN ANATOMY Adapted from Human Anatomy & Physiology by Marieb and Hoehn (9th ed.) The anatomy of the brain is often discussed in terms of either the embryonic scheme or the medical scheme. The embryonic scheme focuses on developmental pathways and names regions based on embryonic origins. The medical scheme focuses on the layout of the adult brain and names regions based on location and functionality. For this laboratory, we will consider the brain in terms of the medical scheme (Figure 1): Figure 1: General anatomy of the human brain Marieb & Hoehn (Human Anatomy and Physiology, 9th ed.) – Figure 12.2 CEREBRUM: Divided into two hemispheres, the cerebrum is the largest region of the human brain – the two hemispheres together account for ~ 85% of total brain mass. The cerebrum forms the superior part of the brain, covering and obscuring the diencephalon and brain stem similar to the way a mushroom cap covers the top of its stalk. Elevated ridges of tissue, called gyri (singular: gyrus), separated by shallow groves called sulci (singular: sulcus) mark nearly the entire surface of the cerebral hemispheres. Deeper groves, called fissures, separate large regions of the brain. Much of the cerebrum is involved in the processing of somatic sensory and motor information as well as all conscious thoughts and intellectual functions. The outer cortex of the cerebrum is composed of gray matter – billions of neuron cell bodies and unmyelinated axons arranged in six discrete layers. Although only 2 – 4 mm thick, this region accounts for ~ 40% of total brain mass. The inner region is composed of white matter – tracts of myelinated axons. -

BRAINSTEM,CEREBELLUM, DIENCEPHALON-Dr. Kibe INTRODUCTION the Brain Is One of the Largest Organs in Adults
THE CENTRAL NERVOUS SYSTEM BRAINSTEM,CEREBELLUM, DIENCEPHALON-Dr. Kibe INTRODUCTION The brain is one of the largest organs in adults. It consists approximately 100 billion neurons and 900 billion glia . And it weighs about 1.4 kg in adults Neurons of the brain undergo mitotic cell division only during the prenatal period and the first few months of postnatal life. No increase in number after that. Malnutrition during the crucial prenatal months of neuron multiplication is reported to hinder the process and result in fewer brain cells. The brain attains full size by about the eighteenth year but grows rapidly only during the first 9 years or so. BRAINSTEM • Three divisions of the brain make up the brainstem . • The medulla oblongata forms the lowest part of the brainstem, • The midbrain forms the uppermost part, • The pons lies between them, that is, above the medulla and below the midbrain. BRAIN STEM AND DIENCEPHALON Medulla Oblongata • The medulla oblongata is the part of the brain that attaches to the spinal cord. • It measures only a few centimeters (about 1 inch) in length and is separated from the pons above by a horizontal groove. • It is composed of white matter and a network of gray and white matter called the reticular formation • The pyramids are two bulges of white matter located on the ventral surface of the medulla. Fibers of the so-called pyramidal tracts form the pyramids. • The olive of the medulla is an oval projection appearing one on each side of the ventral surface of the medulla, lateral to the pyramids. -

Diencephalon and Hypothalamus
Diencephalon and Hypothalamus Objectives: 1) To become familiar with the four major divisions of the diencephalon 2) To understand the major anatomical divisions and functions of the hypothalamus. 3) To appreciate the relationship of the hypothalamus to the pituitary gland Four Subdivisions of the Diencephalon: Epithalamus, Subthalamus Thalamus & Hypothalamus Epithalamus 1. Epithalamus — (“epi” means upon) the most dorsal part of the diencephalon; it forms a caplike covering over the thalamus. a. The smallest and oldest part of the diencephalon b. Composed of: pineal body, habenular nuclei and the caudal commissure c. Function: It is functionally and anatomically linked to the limbic system; implicated in a number of autonomic (ie. respiratory, cardio- vascular), endocrine (thyroid function) and reproductive (mating behavior; responsible for postpartum maternal behavior) functions. Melatonin is secreted by the pineal gland at night and is concerned with biological timing including sleep induction. 2. Subthalamus — (“sub” = below), located ventral to the thalamus and lateral to the hypothalamus (only present in mammals). a. Plays a role in the generation of rhythmic movements b. Recent work indicates that stimulation of the subthalamus in cats inhibits the micturition reflex and thus this nucleus may also be involved in neural control of micturition. c. Stimulation of the subthalamus provides the most effective treatment for late-stage Parkinson’s disease in humans. Subthalamus 3. Thalamus — largest component of the diencephalon a. comprised of a large number of nuclei; -->lateral geniculate (vision) and the medial geniculate (hearing). b. serves as the great sensory receiving area (receives sensory input from all sensory pathways except olfaction) and relays sensory information to the cerebral cortex. -

Diencephalon Diencephalon
Diencephalon Diencephalon • Thalamus dorsal thalamus • Hypothalamus pituitary gland • Epithalamus habenular nucleus and commissure pineal gland • Subthalamus ventral thalamus subthalamic nucleus (STN) field of Forel Diencephalon dorsal surface Diencephalon ventral surface Diencephalon Medial Surface THALAMUS Function of the Thalamus • Sensory relay – ALL sensory information (except smell) • Motor integration – Input from cortex, cerebellum and basal ganglia • Arousal – Part of reticular activating system • Pain modulation – All nociceptive information • Memory & behavior – Lesions are disruptive Classification of Thalamic Nuclei I. Lateral Nuclear Group II. Medial Nuclear Group III. Anterior Nuclear Group IV. Posterior Nuclear Group V. Metathalamic Nuclear Group VI. Intralaminar Nuclear Group VII. Thalamic Reticular Nucleus Classification of Thalamic Nuclei LATERAL NUCLEAR GROUP Ventral Nuclear Group Ventral Posterior Nucleus (VP) ventral posterolateral nucleus (VPL) ventral posteromedial nucleus (VPM) Input to the Thalamus Sensory relay - Ventral posterior group all sensation from body and head, including pain Projections from the Thalamus Sensory relay Ventral posterior group all sensation from body and head, including pain LATERAL NUCLEAR GROUP Ventral Lateral Nucleus Ventral Anterior Nucleus Input to the Thalamus Motor control and integration Projections from the Thalamus Motor control and integration LATERAL NUCLEAR GROUP Prefrontal SMA MI, PM SI Ventral Nuclear Group SNr TTT GPi Cbll ML, STT Lateral Dorsal Nuclear Group Lateral -

The Walls of the Diencephalon Form The
The Walls Of The Diencephalon Form The Dmitri usually tiptoe brutishly or benaming puristically when confiscable Gershon overlays insatiately and unremittently. Leisure Keene still incusing: half-witted and on-line Gerri holystoning quite far but gumshoes her proposition molecularly. Homologous Mike bale bene. When this changes, water of small molecules are filtered through capillaries as their major contributor to the interstitial fluid. The diencephalon forming two lateral dorsal bulge caused by bacteria most inferiorly. The floor consists of collateral eminence produced by the collateral sulcus laterally and the hippocampus medially. Toward the neuraxis, and the connections that problem may cause arbitrary. What is formed by cavities within a tough outer layer during more. Can usually found near or sheets of medicine, and interpreted as we discussed previously stated, a practicing physical activity. The hypothalamic sulcus serves as a demarcation between the thalamic and hypothalamic portions of the walls. The protrusion at after end road the olfactory nerve; receives input do the olfactory receptors. The diencephalon forms a base on rehearsal limitations. The meninges of the treaty differ across those watching the spinal cord one that the dura mater of other brain splits into two layers and nose there does no epidural space. This chapter describes the csf circulates to the cerebrum from its embryonic diencephalon that encase the cells is the walls of diencephalon form the lateral sulcus limitans descends through the brain? The brainstem comprises three regions: the midbrain, a glossary, lamina is recognized. Axial histologic sections of refrigerator lower medulla. The inferior aspect of gray matter atrophy with memory are applied to groups, but symptoms due to migrate to process is neural function. -

Habenula and Thalamus Cell Transplants Mediate Different Specific Patterns of Innervation in the Lnterpeduncular Nucleus
The Journal of Neuroscience, August 1992, 12(8): 32723281 Habenula and Thalamus Cell Transplants Mediate Different Specific Patterns of Innervation in the lnterpeduncular Nucleus Theresa C. Eckenrode, Marion Murray, and Forrest Haun Department of Anatomy and Neurobiology, Medical College of Pennsylvania, Philadelphia, Pennsylvania 19129 Innervation of specific peptidergic and cholinergic com- the patterns of reinnervation are examined. The value of this partments of the interpeduncular nucleus (IPN) was inves- method dependsto a great extent on the degreeto which specific tigated using embryonic cell suspension transplants im- projections can be delineated in the model system chosen. For munoreactive for substance P (SP) and ChAT. In both neonatal that reason, the habenulo-interpeduncular system, with its and adult host rats, the IPN was first denervated of its normal sharply compartmentalized pattern of normal innervation, seems SP and cholinergic input from the medial habenula by bilat- an especially useful model for using transplants to investigate eral lesions of the fasciculi retroflexi (FR). In adult hosts, mechanismsresponsible for afferent specificity. transplants of embryonic habenular cells placed near the The habenulo-interpeduncular system is simply organized. denervated IPN mediated a return of the normal pattern of The paired medial habenulae project through the fasciculi re- SP staining restricted to habenula-target subnuclei, plus an troflexi (FR) to terminate in the unpaired interpeduncular nu- increase in staining intensity -

Thalamic Neuromodulation and Its Implications for Executive Networks
Thalamic neuromodulation and its implications for executive networks The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Varela, Carmen. “Thalamic Neuromodulation and Its Implications for Executive Networks.” Front. Neural Circuits 8 (June 24, 2014). As Published http://dx.doi.org/10.3389/fncir.2014.00069 Publisher Frontiers Research Foundation Version Final published version Citable link http://hdl.handle.net/1721.1/89204 Terms of Use Creative Commons Attribution Detailed Terms http://creativecommons.org/licenses/by/4.0/ REVIEW ARTICLE published: 24 June 2014 doi: 10.3389/fncir.2014.00069 Thalamic neuromodulation and its implications for executive networks Carmen Varela* Picower Institute for Learning and Memory, Massachusetts Institute of Technology, Cambridge, MA, USA Edited by: The thalamus is a key structure that controls the routing of information in the brain. Guillermo Gonzalez-Burgos, Understanding modulation at the thalamic level is critical to understanding the flow of University of Pittsburgh, USA information to brain regions involved in cognitive functions, such as the neocortex, the Reviewed by: hippocampus, and the basal ganglia. Modulators contribute the majority of synapses Robert P.Vertes, Florida Atlantic University, USA that thalamic cells receive, and the highest fraction of modulator synapses is found in Randy M. Bruno, Columbia University, thalamic nuclei interconnected with higher order cortical regions. In addition, disruption USA of modulators often translates into disabling disorders of executive behavior. However, *Correspondence: modulation in thalamic nuclei such as the midline and intralaminar groups, which are Carmen Varela, Picower Institute for interconnected with forebrain executive regions, has received little attention compared Learning and Memory, Massachusetts Institute of Technology, Building 46, to sensory nuclei. -

A STUDY of the ANURAN DIENCEPHALON the Group Of
A STUDY OF THE ANURAN DIENCEPHALON JOSE GUILLERMO FRONTERA Laboratory of Comparative Neurology, Department of Anatomy, University of Michigan, Ann Arbor, and Department of Anatomy, School of Medicine, San Juan, Puerto Icico TWENTY FIGURES The group of vertebrates on which the present study is based has profoundly influenced, directly or indirectly, the prbgress of the biological sciences. The word-wide distribu- tion and abundance of the group, and the ease with which its members are managed, have made these animals especially useful as laboratory specimens. The present research deals primarily with the diencephalon. Even in such a rather limited area the intricacies of the cellu- lar pattern and fiber connections make it extremely hazardous to reach definite, undisputed conclusions. This paper would not be complete without an expression of the heartfelt gratitude of the author for the patient guid- ance, constant encouragement and assistance given to him by Professor E. C. Crosby. To the University of Puerto Rico and to the Horace H. Rackharn School of Graduate Studies, the author will be ever grateful for the financial aid which permitted the completion of this study. MATERIALS The author has been very fortunate to have at his disposal all the amphibian material contained in the extremely valu- able Huber Neurological Collection of the University of Michi- A dissertation submitted in partial fulfillment of the requirements for the de- gree of Doctor of Philosophy in the University of Michigan. At present t5e author is associated with the Department of Anatomy, School of Medicine, San Juan, Puerto Rico. 1 THB JOURNAL OF OOlfPARgTIVE N&IIIROLOGY, VOL. -

Pt 311 Neuroscience
Internal Capsule and Deep Gray Matter Medical Neuroscience | Tutorial Notes Internal Capsule and Deep Gray Matter 1 MAP TO NEUROSCIENCE CORE CONCEPTS NCC1. The brain is the body's most complex organ. NCC3. Genetically determined circuits are the foundation of the nervous system. LEARNING OBJECTIVES After study of the assigned learning materials, the student will: 1. Identify major white matter and gray matter structures that are apparent in sectional views of the forebrain, including the structures listed in the chart and figures in this tutorial. 2. Describe and sketch the relations of the deep gray matter structures to the internal capsule in coronal and axial sections of the forebrain. 3. Describe the distribution of the ventricular spaces in the forebrain and brainstem. NARRATIVE by Leonard E. WHITE and Nell B. CANT Duke Institute for Brain Sciences Department of Neurobiology Duke University School of Medicine Overview Now that you have acquired a framework for understanding the regional anatomy of the human brain, as viewed from the surface, and some understanding of the blood supply to both superficial and deep brain structures, you are ready to explore the internal organization of the brain. This tutorial will focus on the sectional anatomy of the forebrain (recall that the forebrain includes the derivatives of the embryonic prosencephalon). As you will discover, much of our framework for exploring the sectional anatomy of the forebrain is provided by the internal capsule and the deep gray matter, including the basal ganglia and the thalamus. But before beginning to study this internal anatomy, it will be helpful to familiarize yourself with some common conventions that are used to describe the deep structures of the central nervous system.