Resolving Taxonomic Names Using Evidence Extracted from Text
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Florida Native Blackberries Me
The Nature Coastline Newsletter of the Nature Coast Chapter of the Florida Native Plant Society A Message from the President A little bit of Nature eases stress in our everyday life! We all have stress in our lives from not those gardens with stones, ce- one thing or another. We also all ment and alien plants that do not love native plants and know that encourage nature. Even if you our environment is very important spend only a short time outside MARCH-APRIL to us and the future. How can we enjoying your environment and 2020 handle our everyday lives and still breathing the fresh air, you can In this issue: have time to follow the mission of relieve stress. So, the moral of this Meetings/Programs the Florida Native Plant Society of story is that if we plant natives and by Pat Kelly preserving, conserving and restor- encourage wildlife into our back- ing native plants and native plant yards we are also healing our- Calendar communities of Florida? I found selves. In the Spotlight several ways to do this and I hope Thank you for being members of David Barnard you might have your own methods. the Nature Coast Chapter of the One way I found to cope with the Florida Native Plant Society. stress of life and still follow the Lessons from the Landscape by Julie Wert mission is to sit in the backyard with a cup of coffee or a glass of Jonnie Spitler, President Plant Profile: wine! and just enjoy nature around Florida Native Blackberries me. -

Plant Collecting Expedition for Berry Crop Species Through Southeastern
Plant Collecting Expedition for Berry Crop Species through Southeastern and Midwestern United States June and July 2007 Glassy Mountain, South Carolina Participants: Kim E. Hummer, Research Leader, Curator, USDA ARS NCGR 33447 Peoria Road, Corvallis, Oregon 97333-2521 phone 541.738.4201 [email protected] Chad E. Finn, Research Geneticist, USDA ARS HCRL, 3420 NW Orchard Ave., Corvallis, Oregon 97330 phone 541.738.4037 [email protected] Michael Dossett Graduate Student, Oregon State University, Department of Horticulture, Corvallis, OR 97330 phone 541.738.4038 [email protected] Plant Collecting Expedition for Berry Crops through the Southeastern and Midwestern United States, June and July 2007 Table of Contents Table of Contents.................................................................................................................... 2 Acknowledgements:................................................................................................................ 3 Executive Summary................................................................................................................ 4 Part I – Southeastern United States ...................................................................................... 5 Summary.............................................................................................................................. 5 Travelog May-June 2007.................................................................................................... 6 Conclusions for part 1 ..................................................................................................... -
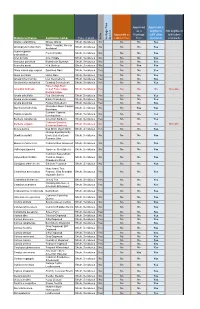
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Complete Iowa Plant Species List
!PLANTCO FLORISTIC QUALITY ASSESSMENT TECHNIQUE: IOWA DATABASE This list has been modified from it's origional version which can be found on the following website: http://www.public.iastate.edu/~herbarium/Cofcons.xls IA CofC SCIENTIFIC NAME COMMON NAME PHYSIOGNOMY W Wet 9 Abies balsamea Balsam fir TREE FACW * ABUTILON THEOPHRASTI Buttonweed A-FORB 4 FACU- 4 Acalypha gracilens Slender three-seeded mercury A-FORB 5 UPL 3 Acalypha ostryifolia Three-seeded mercury A-FORB 5 UPL 6 Acalypha rhomboidea Three-seeded mercury A-FORB 3 FACU 0 Acalypha virginica Three-seeded mercury A-FORB 3 FACU * ACER GINNALA Amur maple TREE 5 UPL 0 Acer negundo Box elder TREE -2 FACW- 5 Acer nigrum Black maple TREE 5 UPL * Acer rubrum Red maple TREE 0 FAC 1 Acer saccharinum Silver maple TREE -3 FACW 5 Acer saccharum Sugar maple TREE 3 FACU 10 Acer spicatum Mountain maple TREE FACU* 0 Achillea millefolium lanulosa Western yarrow P-FORB 3 FACU 10 Aconitum noveboracense Northern wild monkshood P-FORB 8 Acorus calamus Sweetflag P-FORB -5 OBL 7 Actaea pachypoda White baneberry P-FORB 5 UPL 7 Actaea rubra Red baneberry P-FORB 5 UPL 7 Adiantum pedatum Northern maidenhair fern FERN 1 FAC- * ADLUMIA FUNGOSA Allegheny vine B-FORB 5 UPL 10 Adoxa moschatellina Moschatel P-FORB 0 FAC * AEGILOPS CYLINDRICA Goat grass A-GRASS 5 UPL 4 Aesculus glabra Ohio buckeye TREE -1 FAC+ * AESCULUS HIPPOCASTANUM Horse chestnut TREE 5 UPL 10 Agalinis aspera Rough false foxglove A-FORB 5 UPL 10 Agalinis gattingeri Round-stemmed false foxglove A-FORB 5 UPL 8 Agalinis paupercula False foxglove -

North American Species of Rubus L. (Rosaceae) Described from European Botanical Gardens (1789-1823)
adansonia 2021 43 8 DIRECTEUR DE LA PUBLICATION / PUBLICATION DIRECTOR: Bruno David Président du Muséum national d’Histoire naturelle RÉDACTEUR EN CHEF / EDITOR-IN-CHIEF : Thierry Deroin RÉDACTEURS / EDITORS : Porter P. Lowry II ; Zachary S. Rogers ASSISTANT DE RÉDACTION / ASSISTANT EDITOR : Emmanuel Côtez ([email protected]) MISE EN PAGE / PAGE LAYOUT : Emmanuel Côtez COMITÉ SCIENTIFIQUE / SCIENTIFIC BOARD : P. Baas (Nationaal Herbarium Nederland, Wageningen) F. Blasco (CNRS, Toulouse) M. W. Callmander (Conservatoire et Jardin botaniques de la Ville de Genève) J. A. Doyle (University of California, Davis) P. K. Endress (Institute of Systematic Botany, Zürich) P. Feldmann (Cirad, Montpellier) L. Gautier (Conservatoire et Jardins botaniques de la Ville de Genève) F. Ghahremaninejad (Kharazmi University, Téhéran) K. Iwatsuki (Museum of Nature and Human Activities, Hyogo) A. A. Khapugin (Tyumen State University, Russia) K. Kubitzki (Institut für Allgemeine Botanik, Hamburg) J.-Y. Lesouef (Conservatoire botanique de Brest) P. Morat (Muséum national d’Histoire naturelle, Paris) J. Munzinger (Institut de Recherche pour le Développement, Montpellier) S. E. Rakotoarisoa (Millenium Seed Bank, Royal Botanic Gardens Kew, Madagascar Conservation Centre, Antananarivo) É. A. Rakotobe (Centre d’Applications des Recherches pharmaceutiques, Antananarivo) P. H. Raven (Missouri Botanical Garden, St. Louis) G. Tohmé (Conseil national de la Recherche scientifique Liban, Beyrouth) J. G. West (Australian National Herbarium, Canberra) J. R. Wood (Oxford) COUVERTURE -

Shrubs of the Chicago Region
A Selection of Native Shrubs and Noteworthy Non-Native Shrubs 1 WEB VERSION Shrubs of the Chicago Region Volunteer Stewardship Network – Chicago Wilderness Photos by: © Paul Rothrock (Taylor University, IN), © John & Jane Balaban ([email protected]; North Branch Restoration Project), © Kenneth Dritz, © Sue Auerbach, © Melanie Gunn, © Sharon Shattuck, and © William Burger (Field Museum). Produced by: Jennie Kluse © vPlants.org and Sharon Shattuck, with assistance from Ken Klick (Lake County Forest Preserve), Paul Rothrock, Sue Auerbach, John & Jane Balaban, and Laurel Ross. © Environment, Culture and Conservation, The Field Museum, Chicago, IL 60605 USA. [http://www.fmnh.org/temperateguides/]. Chicago Wilderness Guide #5 version 1 (06/2008) EVERGREEN SHRUBS: GROUP 1. LEAVES ARE NEEDLES or SCALES. 1 Juniperus horizontalis TRAILING JUNIPER: 2 Juniperus communis COMMON JUNIPER: Plants have a creeping habit; some leaves are needles but most are Erect shrub or tree (up to 3 m tall); needles whorled on stem; scales with a whitish coat; fruit a bluish-whitish berry-like cone; fruit a bluish or black berry-like cone; grows only in dunes/bluffs male cones on separate plants; grows in sandy soils. bordering Lake Michigan. DECIDUOUS SHRUBS: GROUP 2. LEAVES COMPOUND (more than one leaflet per stalk). STEMS ARMED. 3 Rosa setigera ILLINOIS ROSE: 4 Rosa palustris SWAMP ROSE: Mature plant with long-arching stems; sparse prickles; leaflets Upright shrub; stems very thorny; leaflets 5-7; sepals fall from usually 3, but sometimes 5; styles (female pollen tube) fused into mature fruit; fruit smooth, red berry-like hips; grows in wet and a column; stipules narrow to tip. open ditches, bogs, and swamps. -

Woody Plants
For questions about local plants, call: Natural Resources Coordinator 980-314-1119 www.parkandrec.com WOODY PLANT CHECKLIST Mecklenburg County, NC: 301 species Moschatel Family ☐ Chinese Holly, Burford Holly* ☐ Beaked Hazelnut ☐ Common Elderberry (Ilex cornuta) (Corylus cornuta var. cornuta) (Sambucus canadensis) ☐ Japanese Holly* (Ilex crenata) ☐ American Hop-hornbeam, Ironwood ☐ Maple-leaf Viburnum, Dockmackie ☐ Possum-haw (Ilex decidua var. decidua) (Ostrya virginiana) (Viburnum acerifolium) ☐ Georgia Holly, Chapman’s Holly Bignonia Family ☐ Arrow-wood (Viburnum dentatum) (Ilex longipes) ☐ Cross-vine (Bignonia capreolata) ☐ Southern Wild Raisin, Possumhaw ☐ Mountain Holly (Ilex montana) ☐ Trumpet-creeper (Campsis radicans) (Viburnum nudum) ☐ American Holly, Christmas Holly ☐ Northern Catalpa~ (Catalpa speciosa) ☐ Black Haw (Viburnum prunifolium) (Ilex opaca var. opaca) ☐ Southern Black Haw ☐ Winterberry (Ilex verticillata) Boxwood Family (Viburnum rufidulum) ☐ Yaupon~ (Ilex vomitoria) ☐ Boxwood* (Buxus sempervirens) Agave Family Ginseng Family ☐ Pachysandra, Japanese-spurge* (Pachysandra terminalis) ☐ Rattlesnake-master, Eastern False-aloe ☐ Devil’s-walking-stick, Hercules’s-club (Manfreda virginica) (Aralia spinosa) Cactus Family ☐ Curlyleaf Yucca, Spoonleaf Yucca ☐ Common Ivy, English Ivy* ☐ Prickly-pear (Yucca filamentosa) (Hedera helix var. helix) (Opuntia humifusa var. humifusa) ☐ Weakleaf Yucca (Yucca flaccida) ☐ Ginseng (Panax quinquefolius) Sweet-shrub Family ☐ Mound-lily Yucca~ (Yucca gloriosa) Aster Family ☐ Sweet-shrub, -

Hymenoptera: Apoidea) of the Kitty Todd Preserve, Lucas County, Ohio
The Great Lakes Entomologist Volume 43 Numbers 1 - 4 - 2010 Numbers 1 - 4 - 2010 Article 5 April 2010 Bees (Hymenoptera: Apoidea) of the Kitty Todd Preserve, Lucas County, Ohio Mike Arduser Missouri Department of Conservation Follow this and additional works at: https://scholar.valpo.edu/tgle Part of the Entomology Commons Recommended Citation Arduser, Mike 2010. "Bees (Hymenoptera: Apoidea) of the Kitty Todd Preserve, Lucas County, Ohio," The Great Lakes Entomologist, vol 43 (1) Available at: https://scholar.valpo.edu/tgle/vol43/iss1/5 This Peer-Review Article is brought to you for free and open access by the Department of Biology at ValpoScholar. It has been accepted for inclusion in The Great Lakes Entomologist by an authorized administrator of ValpoScholar. For more information, please contact a ValpoScholar staff member at [email protected]. Arduser: Bees (Hymenoptera: Apoidea) of the Kitty Todd Preserve, Lucas Cou 2010 THE GREAT LAKES ENTOMOLOGIST 45 Bees (Hymenoptera: Apoidea) of the Kitty Todd Preserve, Lucas County, Ohio Mike Arduser 1 Abstract A survey of the bees occurring on the The Nature Conservancy’s Kitty Todd Preserve in the Oak Openings region of Lucas County Ohio was conducted in 2002-2004, using hand-netting techniques. Collecting effort totaled 24.5 hours spread over 11 different days in the three years. All sampling was done in natu- ral communities. One hundred twenty-four species of bees were identified from fifty-one species of flowering plants, including several bee species poorly known or infrequently collected. Comments are provided about the faunistics, bee-plant relationships, oligolecty, management, and natural community dependency of the bees found on the Kitty Todd Preserve. -

Glebe Park Stewardship Plan 2011-2021
DRAFT Glebe Park Stewardship Plan 2011-2021 Produced by Forest Design In association with Glenside Ecological Services Limited June 2011 Glebe Park Stewardship Plan - DRAFT 2011 TABLE OF CONTENTS Introduction ................................................................................................................... 1 Glebe Park Committee objectives ............................................................................................ 1 Recreation ...................................................................................................................... 1 Environment ................................................................................................................... 2 Nature appreciation ........................................................................................................ 2 Scope of Work ......................................................................................................................... 2 General Property Description ....................................................................................... 2 Legal description...................................................................................................................... 2 Activities .................................................................................................................................. 5 History of Glebe Park management ......................................................................................... 5 Surrounding landscape ........................................................................................................... -

Rubus Beamanii, a New Name for Rubus Vagus L.H. Bailey, a Glandular Dewberry Described from Kalamazoo
Widrlechner, M.P. and B.P Riley. 2017. Rubus beamanii , a new name for Rubus vagus L.H. Bailey, a glandular dewberry described from Kalamazoo County, Michigan, and recently discovered in Ohio. Phytoneuron 2017-10: 1–10. Published 1 February 2017. ISSN 2153 733X RUBUS BEAMANII , A NEW NAME FOR RUBUS VAGUS L.H. BAILEY, A GLANDULAR DEWBERRY DESCRIBED FROM KALAMAZOO COUNTY, MICHIGAN, AND RECENTLY DISCOVERED IN OHIO MARK P. WIDRLECHNER Department of Horticulture/Department of Ecology, Evolution, and Organismal Biology Iowa State University Ames, Iowa 50011 BRIAN P. RILEY Ohio Army National Guard Camp Ravenna Joint Military Training Center Newton Falls, Ohio 44444 ABSTRACT The recent discovery of Rubus vagus L.H. Bailey at Camp Ravenna Joint Military Training Center in Portage and Trumbull County, Ohio, has highlighted a seldom-seen species of North American Rubus that, for 70 years, has been illegitimately named. We propose a valid name for it: Rubus beamanii Widrlechner & Riley, nom. nov. , honoring Professor John Beaman (1929-2015). A key to the glandular dewberries of Ohio and surrounding states is included as an aid to distinguishing R. beamanii from the other glandular dewberries encountered in the region. In the early 1940s, the genus Rubus was extensively collected in Kalamazoo Co., Michigan, by Clarence and Florence Hanes and Fred Rapp. Their specimens were shared with Liberty Hyde Bailey for identification in preparation for the publication of “The Flora of Kalamazoo County, Michigan” (Hanes & Hanes 1947). In that book, Bailey described 14 new species of Rubus , including six new members of a group he referred to as sect. -

Checklist of Hinton's Collections of the Flora Of
Acta Botánica Mexicana (1995), 30:41-112 CHECKLIST OF HINTON’S COLLECTIONS OF THE FLORA OF SOUTH-CENTRAL NUEVO LEON AND ADJACENT COAHUILA JAMES HINTON AND GEORGE S. HINTON Rancho Aguililla, Galeana, Nuevo León Apdo. Postal 603, Saltillo, Coahuila, México 25000 ABSTRACT A preliminary checklist of the flora of south-central Nuevo León and part of adjacent Coahuila includes 1955 species, in 713 genera and 130 families. At least 200 species, an estimated 10 percent of the total flora, are endemic to this area, mostly on exposed gypsum and on mountain peaks at the highest elevations. The region of investigation includes the municipios of Allende, Aramberri, Dr. Arroyo, Galeana, Iturbide, Linares, Montemorelos, Rayones, Sabinas Hidalgo, Salinas Victoria, Santa Catarina, Santiago, Villa Aldama, Villa de García and Zaragoza in Nuevo León and Arteaga and Ramos Arizpe in Coahuila, and includes habitats ranging in elevation from 475 to 3700 meters. RESUMEN Se presenta una lista preliminar de la flora del sur y centro de Nuevo León y parte adyacente de Coahuila, la cual incluye 1955 especies en 713 géneros y 130 familias. Por lo menos 200 especies, aproximadamente 10 por ciento de la flora total, son endémicas a esta área, principalmente en suelos yesosos y en cimas de montañas de mayor elevación. La región de investigación abarca los municipios de Allende, Aramberri, Dr. Arroyo, Galeana, Iturbide, Linares, Montemorelos, Rayones, Sabinas Hidalgo, Salinas Victoria, Santa Catarina, Santiago, Villa de Aldama, Villa de García y Zaragoza en Nuevo León, Arteaga y General Cepeda en Coahuila, en un intervalo altitudinal entre 475 y 3700 m. -

Ecological and Land Management Survey and Botanical Inventory for the Nipmuc River Conservation Area in Burrillville, Rhode Island
Ecological and Land Management Survey and Botanical Inventory for the Nipmuc River Conservation Area in Burrillville, Rhode Island Submitted by Jackie Sones, Conservation Biologist Ecological Inventory, Monitoring, & Stewardship Program October 2004 Room 101, Coastal Institute in Kingston 1 Greenhouse Road, URI Kingston, RI 02881 401-874-5817 Table of Contents Page Site location 1 Directions 1 Access/parking 1 Survey dates and observers 1 Property description 1–2 Natural communities 2 Aquatic resources 2–3 Flora 3 Fauna 3–4 Soils 4 Abiotic condition 5 Ecological processes 5 Anthropogenic disturbances 5 Threats (including invasive species) 5–6 Management comments and monitoring needs 6 Adjacent conservation land 6 Inventory needs 6–7 Acknowledgements 7 References 7–8 List of Figures Figure 1. Topographic map of the Nipmuc River Conservation Area Figure 2. Aerial photograph of the Nipmuc River Conservation Area Figure 3. Survey routes covered by RINHS staff in 2004 Figure 4. Features and observations at the Nipmuc River Conservation Area Figure 5. Land use types at the Nipmuc River Conservation Area Figure 6. Locations of rare plants at the Nipmuc River Conservation Area Figure 7. Soil types present at the Nipmuc River Conservation Area Figure 8. Conservation land near the Nipmuc River Conservation Area List of Tables Table 1. List of all plant species recorded at the Nipmuc River Conservation Area in 2004, by life form Table 2. List of bird species recorded along the Nipmuc River Trail Table 3. List of mammal species with the potential to occur at the Nipmuc River Conservation Area Table 4. List of fish species recorded in the Nipmuc River Table 5.