Equine 5Α-Reductase Activity and Expression in Epididymis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Characterization of a Microsomal Retinol Dehydrogenase Gene from Amphioxus: Retinoid Metabolism Before Vertebrates
Chemico-Biological Interactions 130–132 (2001) 359–370 www.elsevier.com/locate/chembiont Characterization of a microsomal retinol dehydrogenase gene from amphioxus: retinoid metabolism before vertebrates Diana Dalfo´, Cristian Can˜estro, Ricard Albalat, Roser Gonza`lez-Duarte * Departament de Gene`tica, Facultat de Biologia, Uni6ersitat de Barcelona, A6. Diagonal, 645, E-08028, Barcelona, Spain Abstract Amphioxus, a member of the subphylum Cephalochordata, is thought to be the closest living relative to vertebrates. Although these animals have a vertebrate-like response to retinoic acid, the pathway of retinoid metabolism remains unknown. Two different enzyme systems — the short chain dehydrogenase/reductases and the cytosolic medium-chain alcohol dehydrogenases (ADHs) — have been postulated in vertebrates. Nevertheless, recent data show that the vertebrate-ADH1 and ADH4 retinol-active forms originated after the divergence of cephalochordates and vertebrates. Moreover, no data has been gathered in support of medium-chain retinol active forms in amphioxus. Then, if the cytosolic ADH system is absent and these animals use retinol, the microsomal retinol dehydrogenases could be involved in retinol oxidation. We have identified the genomic region and cDNA of an amphioxus Rdh gene as a preliminary step for functional characterization. Besides, phyloge- netic analysis supports the ancestral position of amphioxus Rdh in relation to the vertebrate forms. © 2001 Elsevier Science Ireland Ltd. All rights reserved. Keywords: Retinol dehydrogenase; Retinoid metabolism; Amphioxus * Corresponding author. Fax: +34-93-4110969. E-mail address: [email protected] (R. Gonza`lez-Duarte). 0009-2797/01/$ - see front matter © 2001 Elsevier Science Ireland Ltd. All rights reserved. PII: S0009-2797(00)00261-1 360 D. -

Methionine Sulfoxide Reductase a Is a Stereospecific Methionine Oxidase
Methionine sulfoxide reductase A is a stereospecific methionine oxidase Jung Chae Lim, Zheng You, Geumsoo Kim, and Rodney L. Levine1 Laboratory of Biochemistry, National Heart, Lung, and Blood Institute, Bethesda, MD 20892-8012 Edited by Irwin Fridovich, Duke University Medical Center, Durham, NC, and approved May 10, 2011 (received for review February 10, 2011) Methionine sulfoxide reductase A (MsrA) catalyzes the reduction Results of methionine sulfoxide to methionine and is specific for the S epi- Stoichiometry. Branlant and coworkers have studied in careful mer of methionine sulfoxide. The enzyme participates in defense detail the mechanism of the MsrA reaction in bacteria (17, 18). against oxidative stresses by reducing methionine sulfoxide resi- In the absence of reducing agents, each molecule of MsrA dues in proteins back to methionine. Because oxidation of methio- reduces two molecules of MetO. Reduction of the first MetO nine residues is reversible, this covalent modification could also generates a sulfenic acid at the active site cysteine, and it is function as a mechanism for cellular regulation, provided there reduced back to the thiol by a fast reaction, which generates a exists a stereospecific methionine oxidase. We show that MsrA disulfide bond in the resolving domain of the protein. The second itself is a stereospecific methionine oxidase, producing S-methio- MetO is then reduced and again generates a sulfenic acid at the nine sulfoxide as its product. MsrA catalyzes its own autooxidation active site. Because the resolving domain cysteines have already as well as oxidation of free methionine and methionine residues formed a disulfide, no further reaction forms. -

Type of the Paper (Article
Supplementary Material A Proteomics Study on the Mechanism of Nutmeg-induced Hepatotoxicity Wei Xia 1, †, Zhipeng Cao 1, †, Xiaoyu Zhang 1 and Lina Gao 1,* 1 School of Forensic Medicine, China Medical University, Shenyang 110122, P. R. China; lessen- [email protected] (W.X.); [email protected] (Z.C.); [email protected] (X.Z.) † The authors contributed equally to this work. * Correspondence: [email protected] Figure S1. Table S1. Peptide fraction separation liquid chromatography elution gradient table. Time (min) Flow rate (mL/min) Mobile phase A (%) Mobile phase B (%) 0 1 97 3 10 1 95 5 30 1 80 20 48 1 60 40 50 1 50 50 53 1 30 70 54 1 0 100 1 Table 2. Liquid chromatography elution gradient table. Time (min) Flow rate (nL/min) Mobile phase A (%) Mobile phase B (%) 0 600 94 6 2 600 83 17 82 600 60 40 84 600 50 50 85 600 45 55 90 600 0 100 Table S3. The analysis parameter of Proteome Discoverer 2.2. Item Value Type of Quantification Reporter Quantification (TMT) Enzyme Trypsin Max.Missed Cleavage Sites 2 Precursor Mass Tolerance 10 ppm Fragment Mass Tolerance 0.02 Da Dynamic Modification Oxidation/+15.995 Da (M) and TMT /+229.163 Da (K,Y) N-Terminal Modification Acetyl/+42.011 Da (N-Terminal) and TMT /+229.163 Da (N-Terminal) Static Modification Carbamidomethyl/+57.021 Da (C) 2 Table S4. The DEPs between the low-dose group and the control group. Protein Gene Fold Change P value Trend mRNA H2-K1 0.380 0.010 down Glutamine synthetase 0.426 0.022 down Annexin Anxa6 0.447 0.032 down mRNA H2-D1 0.467 0.002 down Ribokinase Rbks 0.487 0.000 -

Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2
biology Article Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2 Chiranan Khantham 1 , Wipawadee Yooin 1,2 , Korawan Sringarm 2,3 , Sarana Rose Sommano 2,4 , Supat Jiranusornkul 1, Francisco David Carmona 5,6 , Wutigri Nimlamool 7 , Pensak Jantrawut 1,2, Pornchai Rachtanapun 8,9 and Warintorn Ruksiriwanich 1,2,8,* 1 Department of Pharmaceutical Sciences, Faculty of Pharmacy, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (C.K.); [email protected] (W.Y.); [email protected] (S.J.); [email protected] (P.J.) 2 Cluster of Research and Development of Pharmaceutical and Natural Products Innovation for Human or Animal, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (K.S.); [email protected] (S.R.S.) 3 Department of Animal and Aquatic Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 4 Plant Bioactive Compound Laboratory (BAC), Department of Plant and Soil Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 5 Departamento de Genética e Instituto de Biotecnología, Universidad de Granada, 18071 Granada, Spain; [email protected] 6 Instituto de Investigación Biosanitaria ibs.GRANADA, 18014 Granada, Spain 7 Citation: Khantham, C.; Yooin, W.; Department of Pharmacology, Faculty of Medicine, Chiang Mai University, Chiang Mai 50200, Thailand; Sringarm, K.; Sommano, S.R.; [email protected] 8 Cluster of Agro Bio-Circular-Green Industry, Faculty of Agro-Industry, Chiang Mai University, Jiranusornkul, S.; Carmona, F.D.; Chiang Mai 50100, Thailand; [email protected] Nimlamool, W.; Jantrawut, P.; 9 School of Agro-Industry, Faculty of Agro-Industry, Chiang Mai University, Chiang Mai 50100, Thailand Rachtanapun, P.; Ruksiriwanich, W. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

A Brief Guide to Enzyme Classification and Nomenclature Rev AM
A Brief Guide to Enzyme Nomenclature and Classification Keith Tipton and Andrew McDonald 1) Introduction NC-IUBMB Enzyme List, or, to give it its full title, “Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes by the Reactions they Catalyse,1 is a functional system, based solely on the substrates transformed and products formed by an enzyme. The basic layout of the classification for each enzyme is described below with some indication of the guidelines followed. More detailed rules for enzyme nomenclature and classification are available online.2 Further details of the principles governing the nomenclature of individual enzyme classes are given in the following sections. 2. Basic Concepts 2.1. EC numbers Enzymes are identified by EC (Enzyme Commission) numbers. These are also valuable for relating the information to other databases. They were divided into 6 major classes according to the type of reaction catalysed and a seventh, the translocases, was added in 2018.3 These are shown in Table 1. Table 1. Enzyme classes Name Reaction catalysed 1 Oxidoreductases *AH2 + B = A +BH2 2 Transferases AX + B = BX + A 3 Hydrolases A-B + H2O = AH + BOH 4 Lyases A=B + X-Y = A-B ç ç X Y 5 Isomerases A = B 6 LiGases †A + B + NTP = A-B + NDP + P (or NMP + PP) 7 Translocases AX + B çç = A + X + ççB (side 1) (side 2) *Where nicotinamide-adenine dinucleotides are the acceptors, NAD+ and NADH + H+ are used, by convention. †NTP = nucleoside triphosphate. The EC number is made up of four components separated by full stops. -

Characterization of Precursor-Dependent Steroidogenesis in Human Prostate Cancer Models
cancers Article Characterization of Precursor-Dependent Steroidogenesis in Human Prostate Cancer Models Subrata Deb 1 , Steven Pham 2, Dong-Sheng Ming 2, Mei Yieng Chin 2, Hans Adomat 2, Antonio Hurtado-Coll 2, Martin E. Gleave 2,3 and Emma S. Tomlinson Guns 2,3,* 1 Department of Pharmaceutical Sciences, College of Pharmacy, Larkin University, Miami, FL 33169, USA; [email protected] 2 The Vancouver Prostate Centre at Vancouver General Hospital, 2660 Oak Street, Vancouver, BC V6H 3Z6, Canada; [email protected] (S.P.); [email protected] (D.-S.M.); [email protected] (M.Y.C.); [email protected] (H.A.); [email protected] (A.H.-C.); [email protected] (M.E.G.) 3 Department of Urologic Sciences, Faculty of Medicine, University of British Columbia, Vancouver, BC V5Z 1M9, Canada * Correspondence: [email protected]; Tel.: +1-604-875-4818 Received: 14 August 2018; Accepted: 17 September 2018; Published: 20 September 2018 Abstract: Castration-resistant prostate tumors acquire the independent capacity to generate androgens by upregulating steroidogenic enzymes or using steroid precursors produced by the adrenal glands for continued growth and sustainability. The formation of steroids was measured by liquid chromatography-mass spectrometry in LNCaP and 22Rv1 prostate cancer cells, and in human prostate tissues, following incubation with steroid precursors (22-OH-cholesterol, pregnenolone, 17-OH-pregnenolone, progesterone, 17-OH-progesterone). Pregnenolone, progesterone, 17-OH-pregnenolone, and 17-OH-progesterone increased C21 steroid (5-pregnan-3,20-dione, 5-pregnan-3,17-diol-20-one, 5-pregnan-3-ol-20-one) formation in the backdoor pathway, and demonstrated a trend of stimulating dihydroepiandrosterone or its precursors in the backdoor pathway in LNCaP and 22Rv1 cells. -

SELENOF) with Retinol Dehydrogenase 11 (RDH11
Tian et al. Nutrition & Metabolism (2018) 15:7 DOI 10.1186/s12986-017-0235-x RESEARCH Open Access The interaction of selenoprotein F (SELENOF) with retinol dehydrogenase 11 (RDH11) implied a role of SELENOF in vitamin A metabolism Jing Tian1* , Jiapan Liu1, Jieqiong Li2, Jingxin Zheng3, Lifang Chen4, Yujuan Wang1, Qiong Liu1 and Jiazuan Ni1 Abstract Background: Selenoprotein F (SELENOF, was named as 15-kDa selenoprotein) has been reported to play important roles in oxidative stress, endoplasmic reticulum (ER) stress and carcinogenesis. However, the biological function of SELENOF is still unclear. Methods: A yeast two-hybrid system was used to screen the interactive protein of SELENOF in a human fetal brain cDNA library. The interaction between SELENOF and interactive protein was validated by fluorescence resonance energy transfer (FRET), co-immunoprecipitation (co-IP) and pull-down assays. The production of retinol was detected by high performance liquid chromatograph (HPLC). Results: Retinol dehydrogenase 11 (RDH11) was found to interact with SELENOF. RDH11 is an enzyme for the reduction of all-trans-retinaldehyde to all-trans-retinol (vitamin A). The production of retinol was decreased by SELENOF overexpression, resulting in more retinaldehyde. Conclusions: SELENOF interacts with RDH11 and blocks its enzyme activity to reduce all-trans-retinaldehyde. Keywords: SELENOF (Seleonoprotein F) , Yeast two hybrid system, Protein-protein interaction, Retinol dehydrogenase 11 (RDH11), Fluorescence resonance energy transfer (FRET), Co-immunoprecipitation (co-IP), Pull- down, Retinol (vitamin a), Retinaldehyde Background SELENOF shows that the protein contains a Selenium (Se) is a necessary trace element for human thioredoxin-like motif. The redox potential of this motif health. -
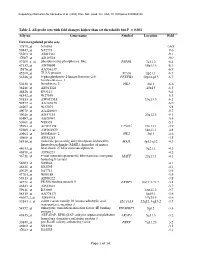
Table 2. All Probe Sets with Fold Changes Higher Than Set Thresholds but P > 0.001 Affy No
Supporting information for Sanoudou et al. (2003) Proc. Natl. Acad. Sci. USA, 10.1073/pnas.0330960100 Table 2. All probe sets with fold changes higher than set thresholds but P > 0.001 Affy no. Gene name Symbol Location Fold Down-regulated probe sets 47879_at N46863 -16.9 59447_at N52773 -9.6 55503_at AI085361 -9.1 47087_at AI310524 -9.0 37209_g_at phosphoserine phosphatase-like PSPHL 7q11.2 -8.4 47137_at AI479899 19p13.3 -8.1 45876_at AA536137 -6.9 45260_at TU3A protein TU3A 3p21.1 -6.7 56246_at 6-phosphofructo-2-kinase/fructose-2,6- PFKFB3 10p14-p15 -6.7 bisphosphatase 3 50230_at hexokinase 2 HK2 2p13 -6.6 38248_at AB011124 20p13 -6.5 44426_at R93141 -6.4 48542_at W27559 -6.3 53813_at AW051518 13q13.3 -6.1 59577_at AA243670 -6.0 46907_at W37075 -5.8 49078_at AA424983 -5.7 49026_at AI357153 20p12.3 -5.4 53487_at AI670947 -5.4 50161_at N39328 -5.1 35994_at AC002398 F25965 19q13.1 -4.9 52285_f_at AW002970 18p11.1 -4.8 40964_at hexokinase 2 HK2 2p13 -4.6 49806_at AI932283 -4.5 58918_at molecule possessing ankyrin repeats induced by MAIL 3p12-q12 -4.3 lipopolysaccharide (MAIL), homolog of mouse 44633_at heat shock 27 kDa associated protein 3q21.1 -4.3 46858_at AI796221 -4.2 36711_at v-maf musculoaponeurotic fibrosarcoma oncogene MAFF 22q13.1 -4.1 homolog F (avian) 54683_at N49844 -4.1 46621_at N32595 -4.1 49629_at N47713 -3.9 47703_at W89189 -3.9 59313_at AI598222 -3.8 34721_at FK506 binding protein 5 FKBP5 6p21.3-21.2 -3.8 46843_at AI632621 -3.7 59611_at R53069 16p11.2 -3.7 58315_at AA778171 3p25.1 -3.6 46607_f_at AI885018 17q25.3 -3.6 33143_s_at solute carrier family 16 (monocarboxylic acid SLC16A3 22q12.3-q13.2 -3.5 transporters), member 3 54152_at eukaryotic translation initiation factor 4E binding EIF4EBP1 8p12 -3.4 protein 1 43935_at ARF-GAP, RHO-GAP, ankyrin repeat and plekstrin ARAP3 5q31.3 -3.2 homology domains-containing protein 3 33849_at pre-B-cell colony-enhancing factor PBEF 7q11.23 -3.2 46902_at N92294 -3.2 47023_at N25555 -3.1 Page 1 of 14 Supporting information for Sanoudou et al. -

Metabolic Enzyme Expression Highlights a Key Role for MTHFD2 and the Mitochondrial Folate Pathway in Cancer
ARTICLE Received 1 Nov 2013 | Accepted 17 Dec 2013 | Published 23 Jan 2014 DOI: 10.1038/ncomms4128 Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer Roland Nilsson1,2,*, Mohit Jain3,4,5,6,*,w, Nikhil Madhusudhan3,4,5, Nina Gustafsson Sheppard1,2, Laura Strittmatter3,4,5, Caroline Kampf7, Jenny Huang8, Anna Asplund7 & Vamsi K. Mootha3,4,5 Metabolic remodeling is now widely regarded as a hallmark of cancer, but it is not clear whether individual metabolic strategies are frequently exploited by many tumours. Here we compare messenger RNA profiles of 1,454 metabolic enzymes across 1,981 tumours spanning 19 cancer types to identify enzymes that are consistently differentially expressed. Our meta- analysis recovers established targets of some of the most widely used chemotherapeutics, including dihydrofolate reductase, thymidylate synthase and ribonucleotide reductase, while also spotlighting new enzymes, such as the mitochondrial proline biosynthetic enzyme PYCR1. The highest scoring pathway is mitochondrial one-carbon metabolism and is centred on MTHFD2. MTHFD2 RNA and protein are markedly elevated in many cancers and correlated with poor survival in breast cancer. MTHFD2 is expressed in the developing embryo, but is absent in most healthy adult tissues, even those that are proliferating. Our study highlights the importance of mitochondrial compartmentalization of one-carbon metabolism in cancer and raises important therapeutic hypotheses. 1 Unit of Computational Medicine, Department of Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 2 Center for Molecular Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 3 Broad Institute, Cambridge, Massachusetts 02142, USA. 4 Department of Systems Biology, Harvard Medical School, Boston, Massachusetts 02115, USA. -

Pyruvate Ferredoxin Oxidoreductase from the Hyperthermophilic
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 9608–9613, September 1997 Biochemistry Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase (archaeayaldehydeydecarboxylationy2-keto acidythiamine pyrophosphate) KESEN MA*, ANDREA HUTCHINS*, SHI-JEAN S. SUNG†, AND MICHAEL W. W. ADAMS*‡ *Center for Metalloenzyme Studies, Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA 30602; and †Institute of Tree Root Biology, U.S. Department of Agriculture–Forest Service, Athens, GA 30602 Communicated by Gregory A. Petsko, Brandeis University, Waltham, MA, June 17, 1997 (received for review June 1, 1996) ABSTRACT Pyruvate ferredoxin oxidoreductase (POR) hyperthermophilic archaea and these are involved in peptide has been previously purified from the hyperthermophilic fermentation. They use 2-ketoglutarate (KGOR) (11), in- archaeon, Pyrococcus furiosus, an organism that grows opti- dolepyruvate (IOR) (12), and 2-ketoisovalerate (VOR) (13) as mally at 100°C by fermenting carbohydrates and peptides. The substrates, and function to oxidatively decarboxylate the 2- enzyme contains thiamine pyrophosphate and catalyzes the keto acids generated by the transamination of glutamate, oxidative decarboxylation of pyruvate to acetyl-CoA and CO2 aromatic amino acids, and branched chain amino acids, re- and reduces P. furiosus ferredoxin. Here we show that this spectively, to the corresponding CoA derivative (13). enzyme also catalyzes the formation of acetaldehyde from The growth of hyperthermophilic archaea such as P. furiosus pyruvate in a CoA-dependent reaction. Desulfocoenzyme A is also unusual in that it is dependent upon tungsten (14), a substituted for CoA showing that the cofactor plays a struc- metal seldom used in biological systems (15). -

A Nitrogenase-Like Methylthio-Alkane Reductase Complex Catalyzes Anaerobic Methane, Ethylene, and Methionine Biosynthesis Justin A
A Nitrogenase-like Methylthio-alkane Reductase Complex Catalyzes Anaerobic Methane, Ethylene, and Methionine Biosynthesis Justin A. North,1 Srividya Murali1* ([email protected]), Adrienne B. Narrowe,3 Weili Xiong,4 Kathryn M. Byerly,1 Sarah J. Young,1 Yasuo Yoshikuni,5 Sean McSweeney,6 Dale Kreitler,6 William R. Cannon,2 Kelly C. Wrighton,3 Robert L. Hettich,4 and F. Robert Tabita1 (former PI, deceased) 1Department of Microbiology, The Ohio State University, Columbus, OH; 2Pacific Northwest National Laboratory, Richland, WA. 3Department of Soil and Crop Sciences, Colorado State University, Fort Collins, CO; 4Chemical Sciences Division, ORNL, Oak Ridge, TN; 5DOE Joint Genome Institute, Berkeley, CA; 6NSLS-II, Brookhaven National Laboratory, Upton, NY. Project Goals: The goal of this project is to identify and characterize the specific enzyme(s) that catalyze anaerobic ethylene synthesis. This is part of a larger project to develop an industrially compatible microbial process to synthesize ethylene in high yields. The specific goals are: 1. Identify the genes and gene products responsible for anaerobic ethylene synthesis. 2. Probe the substrate specificity and metagenomic functional diversity of methylthio-alkane reductases to identify optimal bioproduct generating systems. 3. Characterize the enzymes and the reactions that directly generate anaerobic ethylene. Abstract Text: Our previous work identified a novel anaerobic microbial pathway (DHAP- Ethylene Shunt) [1] that recycled 5’-methylthioadenosine (MTA) back to methionine with stoichiometric amounts of ethylene produced as a surprising side-product. MTA is a metabolic byproduct of methionine utilization in a multitude of cellular processes. The initial steps of the DHAP-ethylene sequentially converts MTA to dihydroxyacetone phosphate (DHAP) and ethylene precursor (2-methylthio)ethanol (Fig.